Development and application method of solanum melongena L. EST-SSR marker
An application method, eggplant technology, applied in the field of breeding, can solve the problems of low marker polymorphism, limited sequence information, weak research foundation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment 1
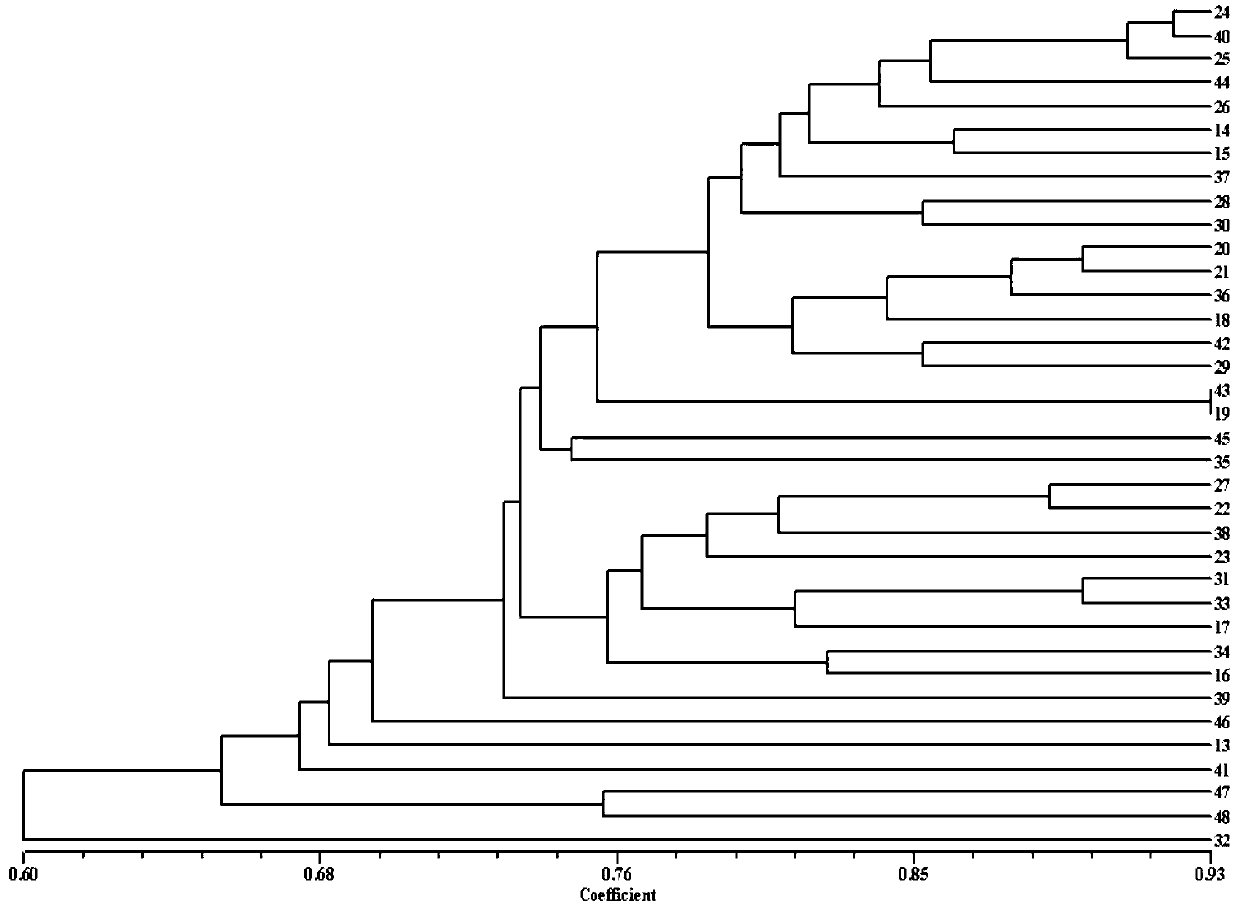
[0072] 1. Experimental material selection: Eggplant materials numbered 1-48 as shown in Table 1 were taken and planted in the experimental greenhouse.
[0073] 2. Get the tender green leaves from the field material and extract its total DNA. The specific steps are as follows:
[0074] (1) Put 100-200mg frozen leaves or fresh leaves of tender green leaves into a pre-cooled mortar, add liquid nitrogen to grind. Transfer the ground powder into a 2ml centrifuge tube, add 800ul freshly prepared extraction buffer (0.35M Glucose, 0.1M Tris.HCl, 5mM Na.EDTA, 2%PVP, 1%(V / V)β-Me, dd Water), vortexed and kept in ice bath for 10 min. Centrifuge at 10000rpm for 15min (4°C), discard the supernatant;
[0075] (2) Add 700ul 65°C preheated lysis buffer (1.4M NaCl, 0.1M Tris.HCl, 20mM Na.EDTA, 2%CTAB, 2%PVP, 1%(V / V)β-Me, ddWater), and use a toothpick to loosen, vortex to mix, 65 ℃ water bath for 30min;
[0076] (3) Add 750ul of chloroform: isoamyl alcohol (24:1) mixture, invert more than 50...
specific Embodiment 2
[0096] 1. The eggplant parents numbered 44 and 41, F 1 and its F 2 Group planted in greenhouses;
[0097] 2. Take tender green leaves from the field materials and extract their total DNA.
[0098] 3. Use the developed primers for analysis: the specific steps are as follows:
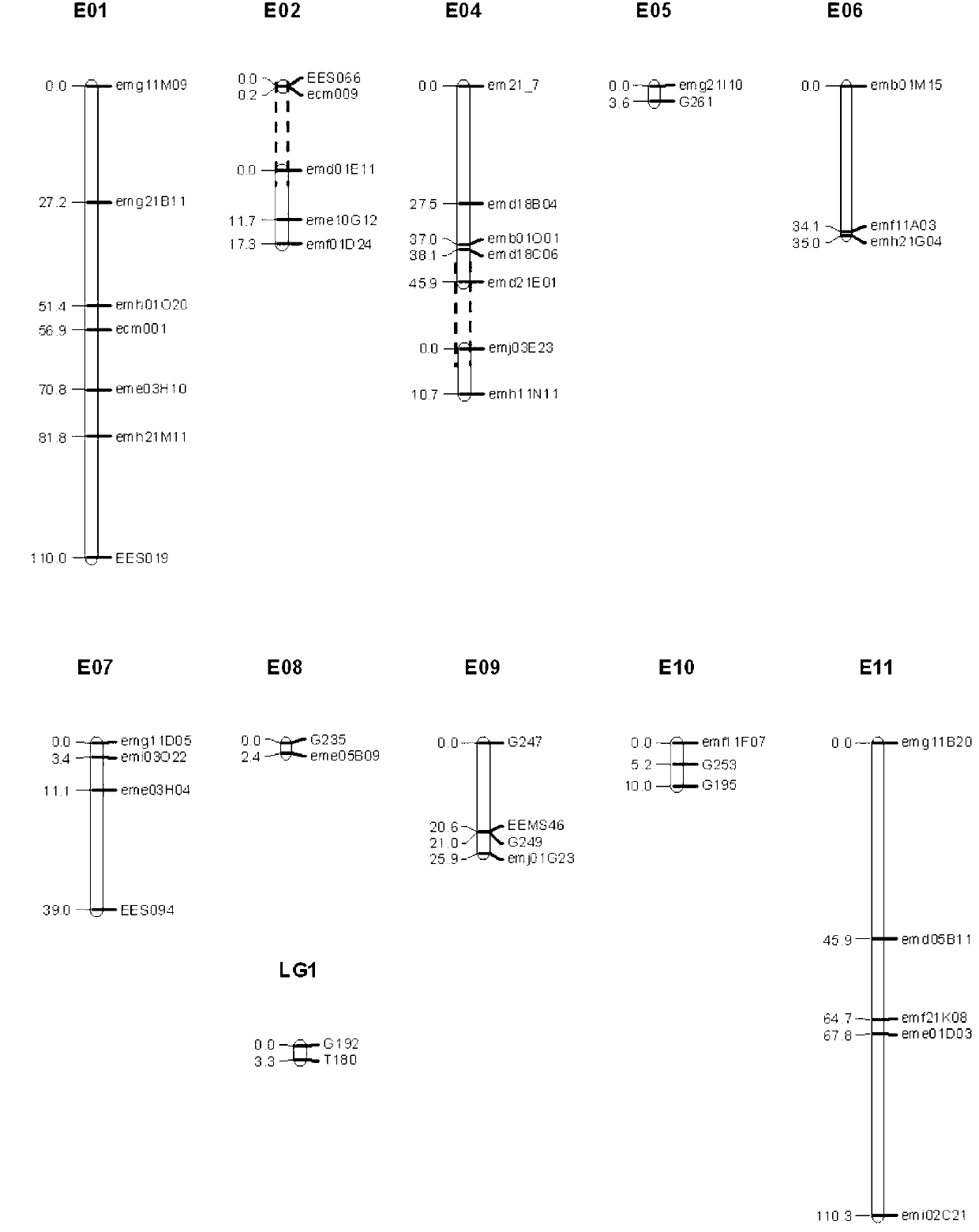
[0099] 3.1 Use the 43 pairs of markers developed by this marker to analyze;
[0100] The PCR reaction system of SSR is: the total volume of the system is 10 μL, the template DNA is about 20 ng., the front primer and the back primer are respectively 0.1 μM, 2.5 mM MgCl 2 , 0.2mM dNTPs, 1×Taq buffer and 1U Taq DNA Taq polymerase (Shanghai Promega). The PCR reaction was performed on a 96-well PCR machine (Eppendorf AG6321, Eppendorf). The reaction program was: pre-denaturation at 94°C for 3 min; denaturation at 94°C for 0.5 min, renaturation at 55°C for 1 min, extension at 72°C for 1 min, 30 cycles; extension at 72°C for 5 min and storage at 4°C.
[0101] The amplified product undergoes non-denaturin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com