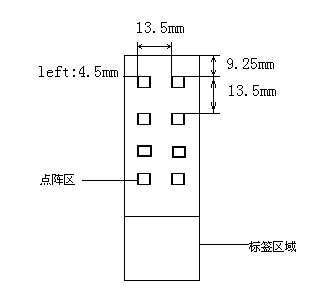
Genetic typing chip of 10 common pathogenic legionella and detection kit
A gene chip and Legionella technology, applied in the field of gene chips and detection kits containing the chip, can solve the problems of low resolution, inability to distinguish species, inability to distinguish Escherichia coli and Shigella, etc., and achieve repeatability Strong, accurate and easy-to-use effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0086] Example 1 Design and preparation of probes
[0087] 1. Sequence acquisition:
[0088] (1) Obtaining the ITS sequence of Legionella longbeach: the ITS sequence of Legionella longbeach was downloaded from the GenBank public database;
[0089] (2) Legionella pneumophila, Legionella anserii, Legionella bozemannii, Legionella Dumov, Legionella feifei, Legionella golemannii, Legionella jordan river, Legionella mirini, Legionella mei The ITS sequence of was deciphered by our laboratory;
[0090] 2. Example of probe design:
[0091] Probes for Legionella anseri: Import the ITS sequence of Legionella anseri into Glustal X software, select a representative sequence and perform Blastn comparison in the public data NCBI to determine whether it can be used as a specific target and the position of the specific target. Import the sequences into OligoArray 2.0 software. Run the program to design probes online.
[0092] 3. Probe synthesis: Extend the 5' end of the probe sequence in...
Embodiment 2
[0099] Example 2 Design and preparation of primers
[0100] 1. Example of primer design:
[0101] Because the 16s and 23s sequences are conserved among different Legionella species, we can ensure that all Legionella can be amplified by designing universal primers at the end of 16s and the beginning of 23s. Taking upstream primers as an example, first put the 16s sequences of different bacteria into MEGA software for analysis, and select the most versatile fragments to import into Primer Primer 5.0 software, set the length to 70bp-10bp, and G+C% value to 40%-60 %, Hairpin: NONE, Dimer: NONE, False Priming: NONE, Cross Dimer: NONE.
[0102] The design method of the downstream primer is the same as that of the above-mentioned probe primer, and the design parameters used are also the same.
[0103] 2. Primer synthesis: commission the primer sequences in Table 2 to be synthesized (PAGE purified) by Probe Synthesis Co., Ltd.
[0104] SEQ ID serial number Sequence (5...
Embodiment 3
[0106] Specific identification of a genotyping array for the detection of important pathogenic Legionella species
[0107] The specificity experiment of gene chip of the present invention:
[0108] Specificity means that a probe can only hybridize with the target gene of its corresponding bacterial species and cannot hybridize with gene fragments of other bacterial species. This requires us not only to carry out a large number of bioinformatics analysis, but the probes analyzed by bioinformatics will also have non-specific results or unstable hybridization signals. Therefore, probe-specific screening still needs a large number of experiments to verify. In particular, some probes used for high-throughput detection must be verified by hybridization with a large number of close and distant bacteria.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com