Degradation of PA200 and acetylation-mediated core histones through proteasome
An acetylation and histone technology, applied in the fields of peptide/protein components, hydrolase, drug combination, etc., can solve the problems of unclear acetylation spermatogenesis and DNA repair
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
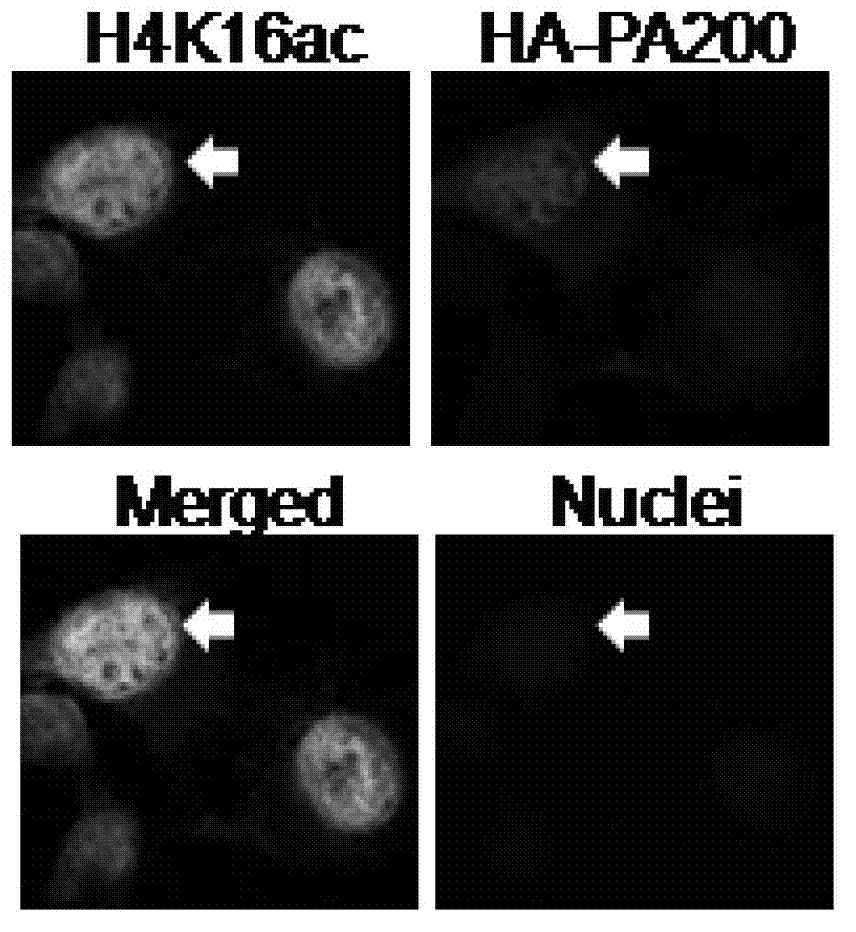
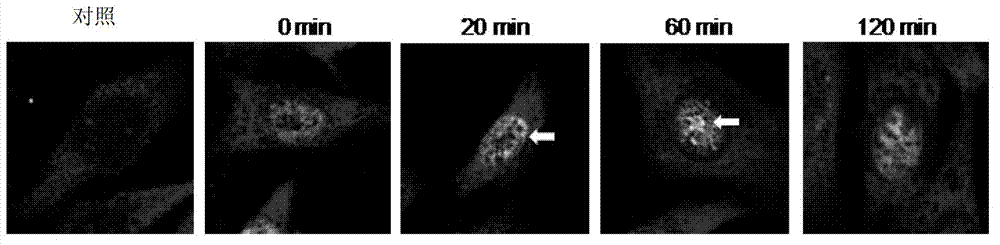
[0063] Example 1. PA200 recognizes and binds to acetylated histones
[0064] 1. Co-localization of PA200 and H2BK5ac
[0065] Preparation method of mouse H2BK5ac antibody: H2BK5ac synthesized by GenScript (Nanjing, China) (H2BK5ac represents histone H2B acetylated at the K5 site, with a biotin label, specifically: PEPAKacSAPAPKKGSKKAVTKA-Biotin) immunized BALB / C Mice, collect serum.
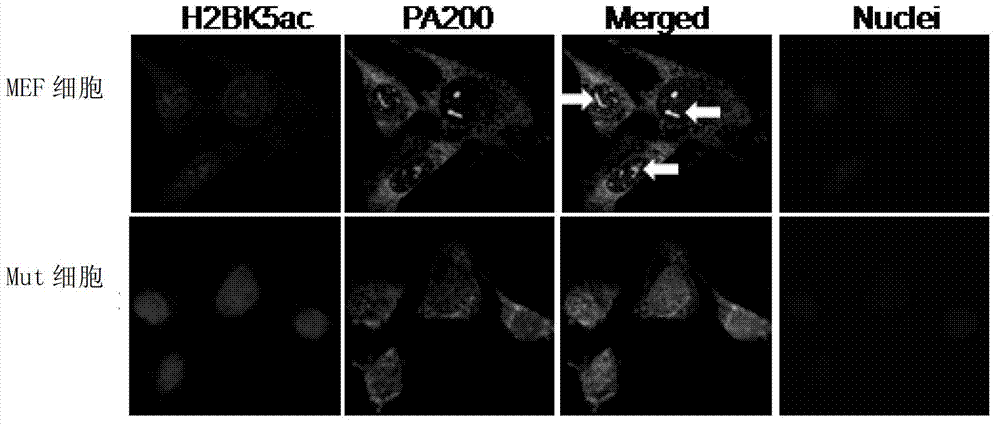
[0066] Take MEF cells or Mut cells and use mouse H2BK5ac antibody (the corresponding secondary antibody is goat anti-mouse-Dylight594, red) and rabbit PA200 antibody (the corresponding secondary antibody is goat anti-rabbit-FITC, green) for immunofluorescence Stain, DAPI stains the nucleus.
[0067] See you in the photo figure 1 (The arrow indicates a trajectory for each cell). In MEF cells, PA200 antibody recognizes punctate structures in the nucleus, and H2BK5ac also appears in this area. The staining intensity of H2BK5ac in Mut cells was significantly greater than that of MEF cells. The results sh...
Embodiment 2
[0112] Example 2. Identification of two different types of proteasomes in mammalian testes
[0113] Histones are lost in large amounts in spermatogenic cells. In order to find the potential mechanism of histone degradation, this example detects whether mammalian spermatogenic cells have a unique proteasome form.
[0114] 1. Extract the total protein of bovine skeletal muscle (muscle), which is total muscle protein; extract the total protein of bovine testicular seminiferous tubules, which is total spermatogenic protein.
[0115] 2. Perform non-denaturing gel electrophoresis on total muscle protein and total spermatogenic protein, and incubate with fluorescent peptide substrate (succinyl LLVY-7-amino-4-methylcoumarin) (for co-incubation, set two treatments, one Add SDS with a final concentration of 0.02% during the incubation, and do not add SDS during the other incubation; after shaking at room temperature for 15 minutes, observe the proteasome band under UV light.
[0116] See the re...
Embodiment 3
[0132] Example 3. Specific subunits and activities in the spermatogenic proteasome
[0133] 1. C2C12 myoblasts, TM3 Leydig cells, TM4 Sertoli cells, mouse GC-1spg type B spermatogonia (GC1 cells for short), mouse GC-2spd(ts) spermatocytes (GC2 cells for short) ), sperm (sperm), testis tissue (testis), spleen tissue (spleen), respectively extract total protein and perform SDS-PAGE electrophoresis and immunoblotting analysis. Each of the above cells were purchased from the American Model Culture Collection Bank. All the above tissues are the tissues of BALB / C mice. The primary antibodies used in western blotting were rat anti-α4s antibody, rat anti-α4 antibody, β1i antibody, β1 antibody, β2i antibody, β2 antibody, β5i antibody, β5 antibody, PA28α antibody, PA200 antibody, Rpt4 antibody, Rpn7 antibody and GAPDH antibody.
[0134] See the result Figure 8 A. α4s and β2i can only be detected in Sertoli cells and spermatocytes, while β5i, PA200, 19S subunits (such as Rpt2, Rpt4 and R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com