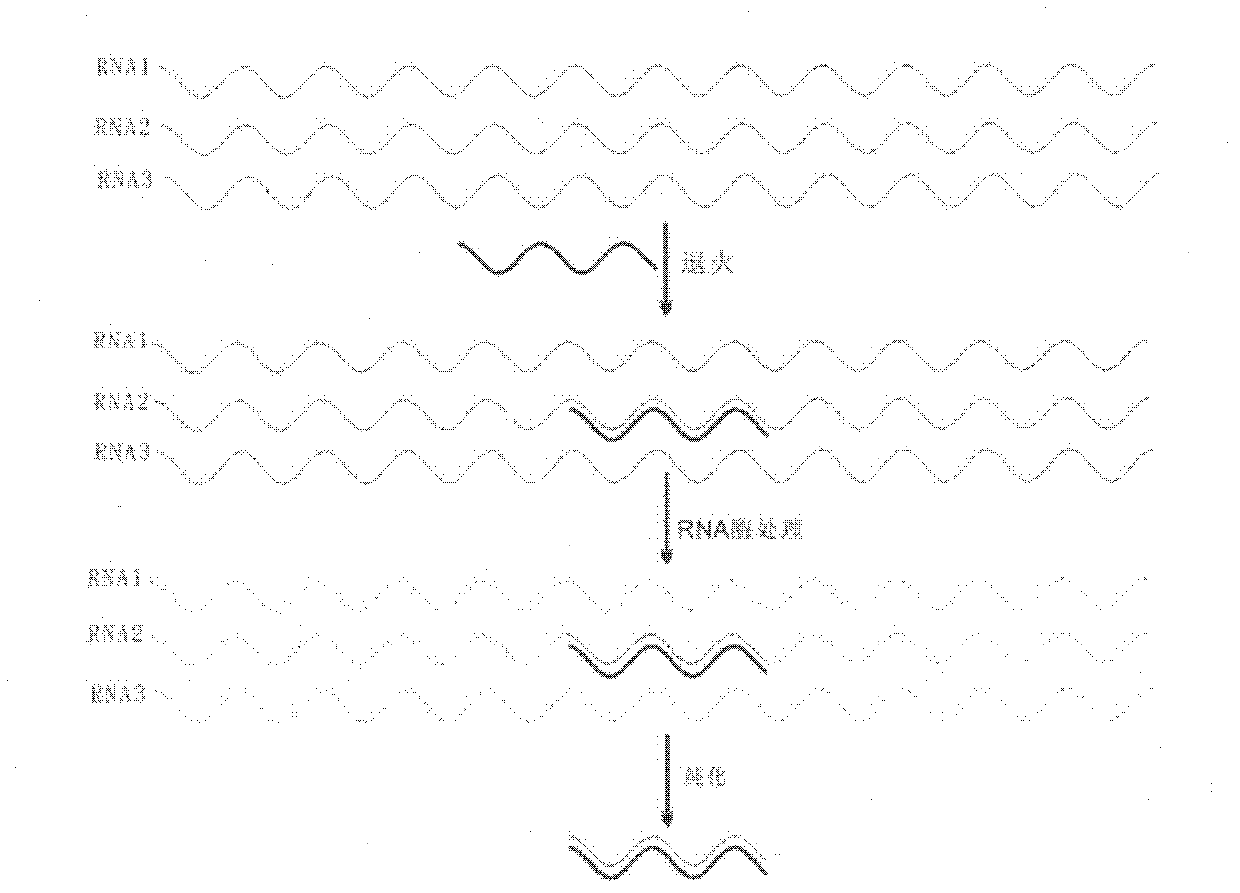
Method for separating low copy genes
A separation method and low-copy technology, applied in the field of gene separation, to achieve the effects of high detection success rate, simple operation, and improved detection efficiency and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
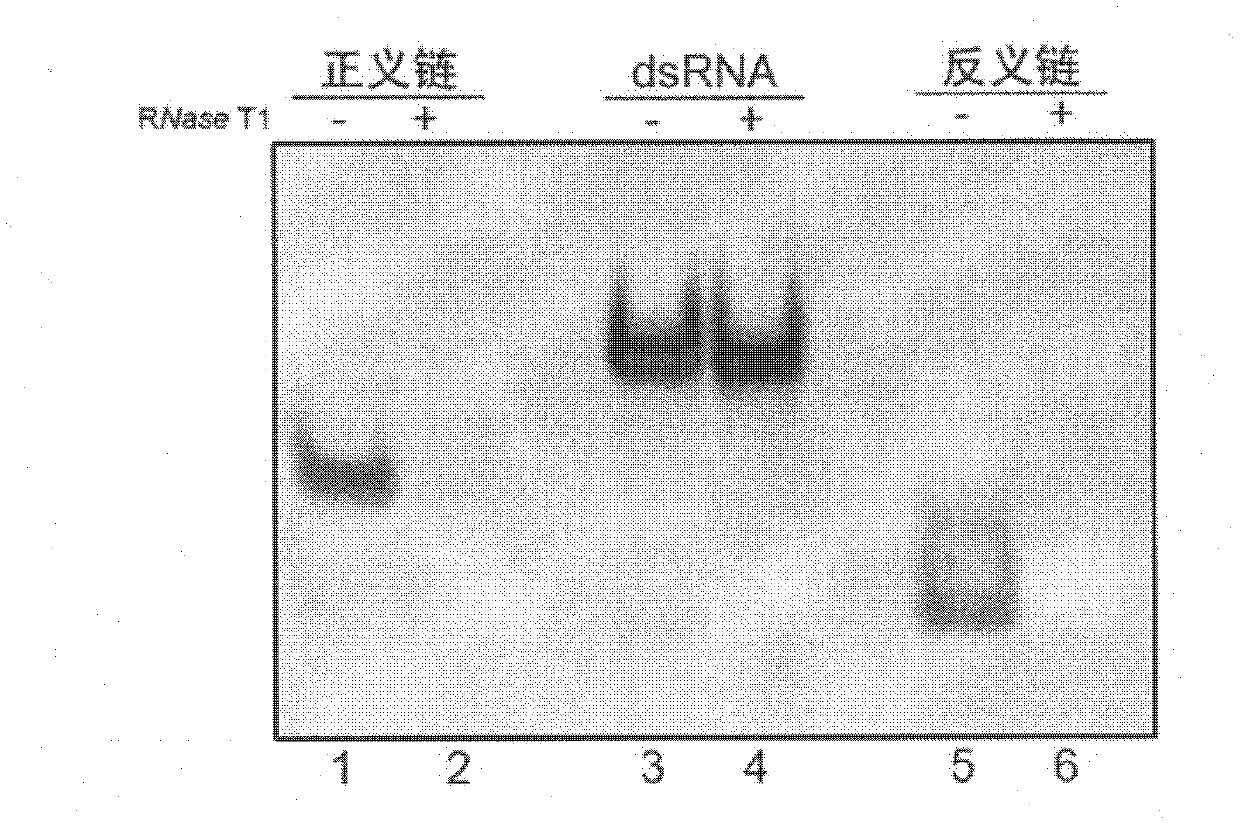
[0059] Example 1: RNase T1 specific digestion of single-stranded RNA experiment
[0060] 1 Main reagents, materials and instruments
[0061] 1.1 ssRNA: si-NC sense strand and antisense strand, synthesized by Biomaike Biotechnology Co., Ltd.
[0062] si-NC sequence (5'-3') (the terminal "dTdT" is a DNA base):
[0063] Sense strand siNC F: UUCUCCGAACGUGUCACGUdTdT (SEQ ID NO: 1);
[0064] Antisense strand siNC R: ACGUGACACGUUCGGAGAAdTdT (SEQ ID NO: 2).
[0065] 1.2 Reagent: RNase T1 (Fermentas company), 40% polyacrylamide, 10× Tris-borate-EDTA (TBE) buffer (containing 890mM Tris-H 3 BO 3 and 20mM EDTA, pH=8.3), 10% (W / V) ammonium persulfate (APS), tetramethyldiethylamine (TEMED), DEPC-H 2 O (all reagents are autoclaved), etc.
[0066] 1.3 Instruments: polyacrylamide gel electrophoresis system (Bio-Rad Company); gel imaging system (Biosens Company), etc.
[0067] 2 RNase T1 digestion reaction
[0068] Dilute the sense strand and antisense strand of si-NC to a certain conce...
Embodiment 2
[0076] Example 2: RNase T1 has no sequence-dependent experiment on the effect of dsRNA
[0077] 1 Main reagents, materials and instruments
[0078] 1.1 The RNA sequence in the following table is taken as an example, synthesized by Biomaike Biotechnology Co., Ltd.
[0079]
[0080] 1.2 Reagent: RNase T1 (Fermentas Company), 40% polyacrylamide, 10× TBE buffer (containing 890mM Tris-H 3 BO 3 and 20mM EDTA, pH=8.3), 10% (W / V) ammonium persulfate, tetramethyldiethylamine, DEPC-H 2 O (all reagents are autoclaved), etc.
[0081] 1.3 Instruments: polyacrylamide gel electrophoresis system (Bio-Rad Company); gel imaging system (Biosens Company), etc.
[0082] 2 RNase T1 digestion reaction
[0083] Dilute the sense and antisense strands of the RNA to a certain concentration, and take a part to anneal into double strands. Take 0.1OD ssRNA and dsRNA and react with 100U RNase T1 at 37°C for 10min.
[0084] 3 Polyacrylamide gel electrophoresis detection
[0085] 3. Preparation of ...
Embodiment 3
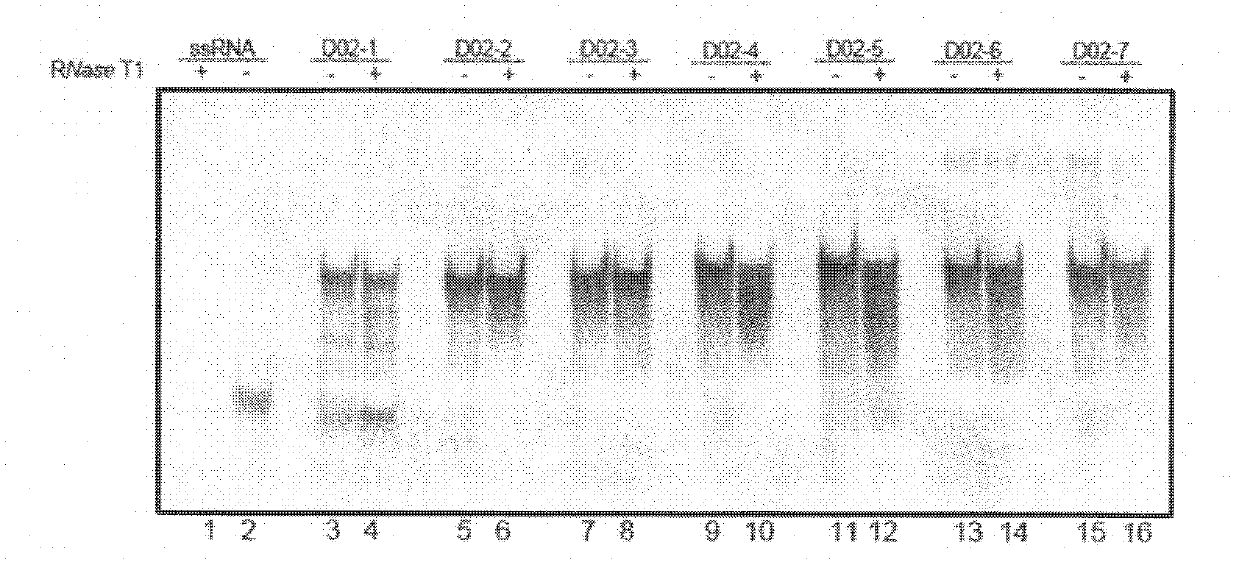
[0091]Embodiment 3: Experiment of the effect of RNase T1 on long single-stranded RNA
[0092] 1 Main reagents, materials and instruments
[0093] 1.1 Reagents: RNase T1 (Fermentas Company), T7 in vitro transcription kit (Biomark Biotechnology Co., Ltd.), 40% polyacrylamide, 10×TBE buffer (containing 890mM Tris-H 3 BO 3 and 20mM EDTA, pH=8.3), 10% (W / V) ammonium persulfate, tetramethyldiethylamine, DEPC-H 2 O (all reagents are autoclaved), etc.
[0094] 1.2 Instruments: polyacrylamide gel electrophoresis system (Bio-Rad Company); gel imaging system (Biosens Company), etc.
[0095] 2 Preparation of long single-stranded RNA (Long single-strand RNA, LRNA)
[0096] Using the CDS sequence of the gene bank number NM 198159 in the NCBI database of the United States as the experimental template, LRNA was synthesized by T7 in vitro transcription method, with a length of 1563nt.
[0097] The primers used for in vitro transcription template preparation are:
[0098] Upstream primer ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com