Beta-agarase and application thereof
An agarase, agar technology, applied in the application, enzyme, hydrolase and other directions, can solve problems such as oligosaccharides that cannot produce a single degree of polymerization, and achieve the effect of good biocatalytic activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1 Cloning of β-agarase gene agWH50C
[0019] The β-agarase gene agWH50C in Agarophage agarophora WH0801 (NRRLB-59247(T) or CGMCC1.10131(T)) was cloned by degenerate primer PCR and nested PCR. Specific operation: Cultivate Agarophore WH0801 in 2216E seawater medium until the end of the logarithm, and extract the DNA genome. Using degenerate primers (5'-HTRCCNAAYCAYHTRTAYHTRGGN-3'; 5'-VACRAARCCVACRTTRTARTTTTC-3'), using the genome as a template, the conditions are: 94°C for 5 minutes, then 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 3 minutes Perform 30 cycles in 3 steps, and perform PCR to obtain a fragment of the β-agarase gene, then design primers based on the end sequence of the fragment, and perform three rounds of nested PCR to recover PCR products with a size of 500Kb-2000Kb Fragments (using the chromosome walking kit from Takara Company). The PCR products in each step were ligated to the T-A cloning vector and then transformed into DH5α and pi...
Embodiment 2
[0020] Example 2 Construction of expression plasmid pETC containing β-agarase gene agWH50C
[0021] According to the obtained full-length sequence of the β-agarase gene, the upstream and downstream primers of the gene were designed (see Example 1), and the genome of Agarophore chrysogenum WH0801 was used as a template to obtain β-agarase by PCR amplification The full-length sequence of the gluease gene. Conditions were 2 minutes at 94°C, followed by 30 seconds at 94°C, 30 seconds at 55°C, 3 minutes at 72°C, 30 cycles and a final extension at 72°C for 10 minutes. Agarose gel electrophoresis showed that there was an obvious single band at the position of 2.8kb, and the single band was recovered by excising the gel. The sequencing results showed that the sequence obtained by splicing in Example 1 was correct. The recovered band was double-digested with Xho1 and Sac1, and the expression vector pET21a was also double-digested with Xho1 and Sac1, and then both the PCR product and t...
Embodiment 3
[0022] Example 3 Construction of engineering bacteria pETC / BL21 highly expressing β-agarase gene agWH50C
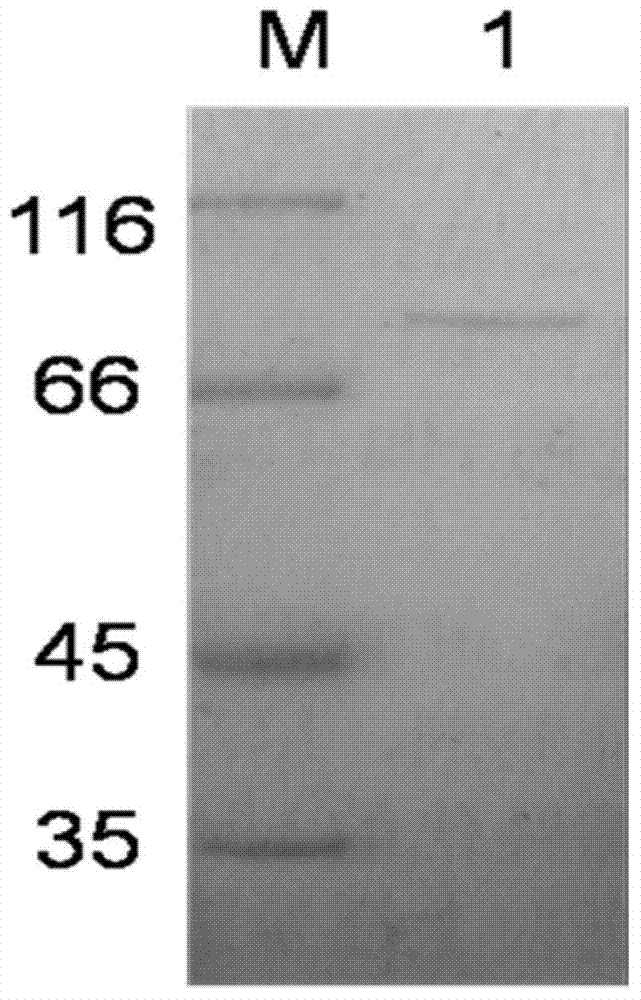
[0023] The expression vector plasmid pETC was transformed into BL21(DE3) according to standard calcium chloride heat shock transformation, and positive transformants with ampicillin resistance were screened. The plasmid was extracted by standard alkaline lysis method, and the plasmid was double digested with Xho1 and Sac1. The gel electrophoresis bands showed two clear bands of 2.8Kb and 5.3Kb, which corresponded to the full length of the β-agarase gene and the length of the plasmid. It proved that the expression vector plasmid pETC containing β-agarase gene had been transferred into BL21 (DE3).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com