Using amRNAs to obtain a safe and alternative way to kill transgenic rice
A technology of genetically modified rice and extermination, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Embodiment 1, Stem-loop backbone endogenous miRNA selection:
[0047] Stem-loop backbone endogenous miRNAs were selected from all rice miRNA precursors present in miR / Rfam by 2 criteria, which are as follows:
[0048] Criterion 1. The predicted secondary structure can form a short, simple hairpin stem-loop structure, and at the same time can form a complete miRNA-miRNA* matching endogenous miRNA precursor without insertion or deletion;
[0049] Criterion 2. The endogenous miRNA precursor sequence already exists in the EST database and is supported by corresponding literature.
[0050] osa-MIR528 (acquisition number: MI0003201; corresponding literature: NormanWarthmannetal, PLoSONE, 2008, 3(3).) meets the above two criteria.
[0051] The precursor of osa-MIR528 (acquisition number: MI0003201) can form a short stem-loop structure, and it exists and expresses in rice roots, leaves and other tissues (LiuBetal, PlantPhysiol2005, 139:296–305); via NCBI There are 3 EST seque...
Embodiment 2
[0053] Example 2. Artificial amiRNA selection:
[0054] WMD2 (http: / / wmd2.weigelworld.org) currently provides miRNA sequences of 21 bases in 38 plants including rice. It (WMD2) provides specific and effective candidate miRNAs through two-step screening. First, the The database software will regard all sequences whose 10th base in the input sequence is "A" or "U" and whose 5' end is unstable (5' high AU content, 3' high GC content) with a length of 21 bases as candidates miRNA, then replace the first base of all candidate miRNAs with "U", and then continuously replace 13-15 and 17-21 bases of candidate miRNAs based on specific species genes or sequences, which will be in the "seed region" (2-12 Base region) has no more than 1 base mismatch with the target sequence, and candidate miRNAs with 1 or 2 base mismatches at other positions are returned to the user as the final miRNA (SchwabRetal, PlantCell, 2006, 18:1121 –1133; Ossowski Setal, Plant J, 2008, 53:674–690; Schwab Retal, ...
Embodiment 3
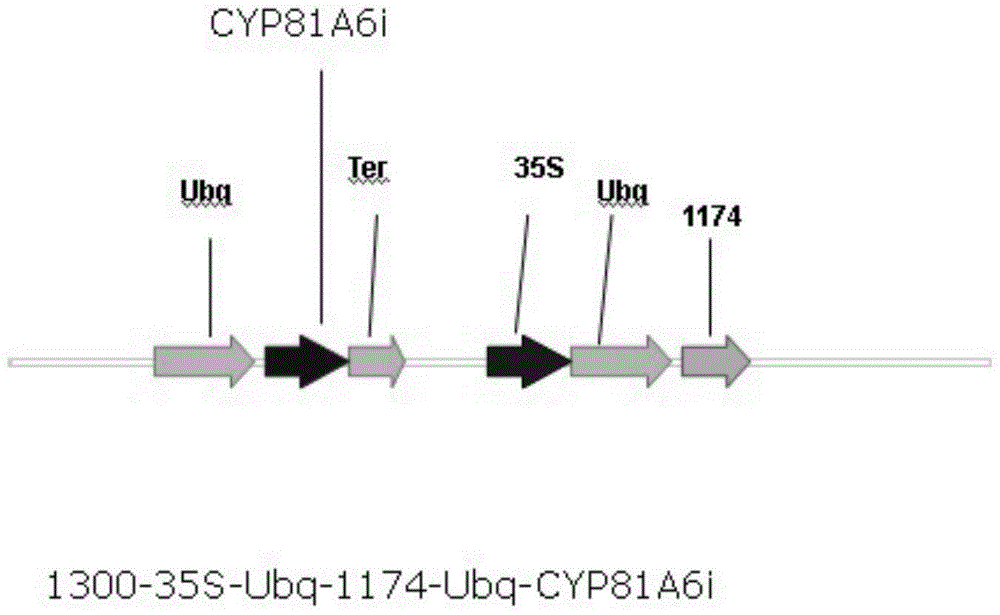
[0060] Example 3, Construction of Rice Expression Vector 1300-35S-Ubq-1174-Ubq-CYP81A6i
[0061] Using primer miR1-F:AG TCAATACATGAGTTCGTCCGG CAGGAGATTCAGTTTGA (the underline represents the replacement base);
[0062] miR1-R:TG CCGGACGAACTCATGTATTGA CTGCTGCTGCTACAGCC (the underline represents the replacement base);
[0063] miR11-F: CT CCGGAgGAAgTCATGTATTGA TTCCTGCTGCTAGGCTG (underlines represent replacement bases, lowercase letters represent mismatched bases);
[0064] miR11-R:AA TCAATACATGAcTTCcTCCGG AGAGAGGCAAAAGTGAA (underlines represent replacement bases, lowercase letters represent mismatched bases);
[0065] Replace the 21bp base sequence in the 245bp endogenous miRNAosa-MIR528 (Note: miR1-R contains the amiRNA sense strand sequence, miR1-F contains the amiRNA antisense strand sequence, miR11-F contains the amiRNA* sense strand sequence, and miR11-R contains the amiRNA* sense strand sequence amiRNA* sense strand sequence).
[0066] Remarks: In SEQ ID NO: 1, the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com