Enrichment and detection method of target gene fragment
A technology of target genes and fragments, applied in biochemical equipment and methods, microbial determination/testing, DNA preparation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0144] Example 1: Using the method of the present invention to enrich and detect the KRAS gene in cells
[0145] In this example, the KRAS gene of the MDA-MB-231 cell line was enriched and detected.
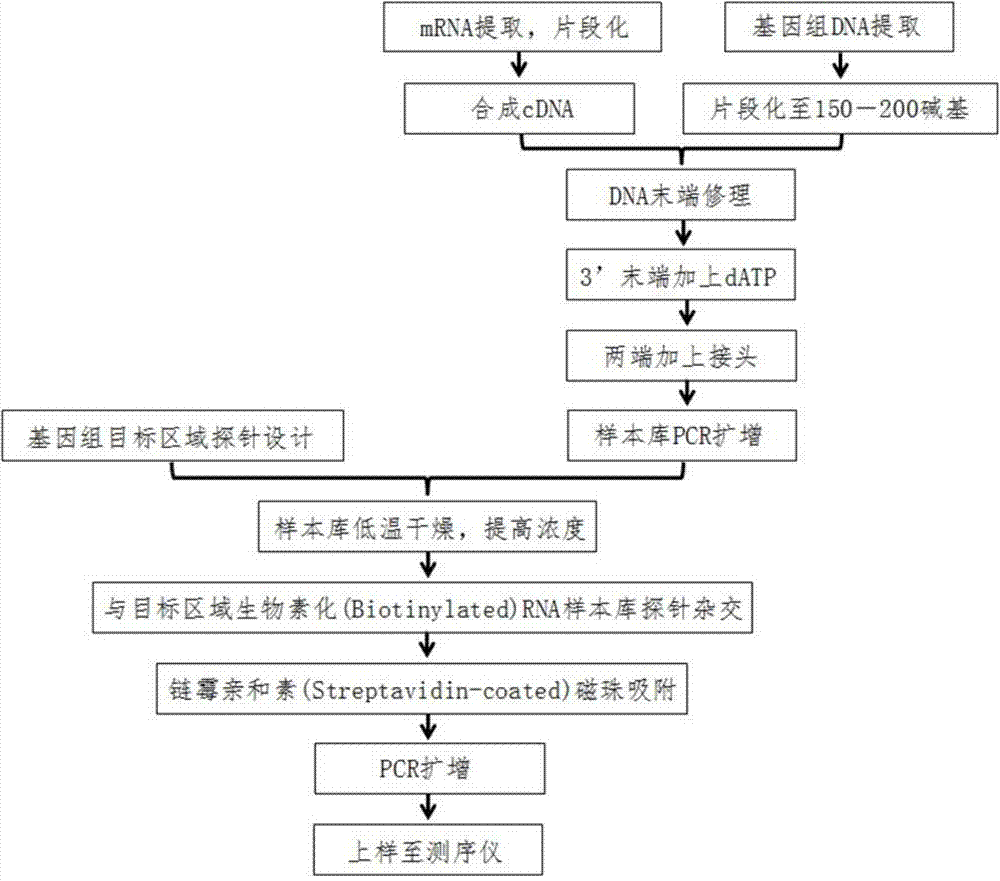
[0146] 1. Prepare the DNA sample bank of the MDA-MB-231 cell line to be tested
[0147] 1. Extract the whole genome DNA of the MDA-MB-231 cell line, and then fragment it (the DNA sample bank obtained in this way is called "DNA sample bank derived from the whole genome")
[0148] 1.1 DNA extraction
[0149] Using Qiagen Blood&Tissue DNeasy kit (article number: 69506), whole genome DNA was extracted from the MDA-MB-231 cell line (human breast cancer cell line, from the ATCC cell bank, article number: HTB-26). Follow the instructions in the manual.
[0150] Use a spectrophotometer and a gel electrophoresis system to detect the quality and concentration of DNA. The 260nm absorbance of dsDNA is greater than 0.05, and the absorbance ratio of A260 / A280 is between 1.8 and 2.
[0151] 1.2 DNA frag...
Embodiment 2
[0229] Example 2: Using the method of the present invention to enrich and detect the KRAS gene in cells
[0230] In this example, the KRAS gene of the HCT-116 cell line was enriched and detected.
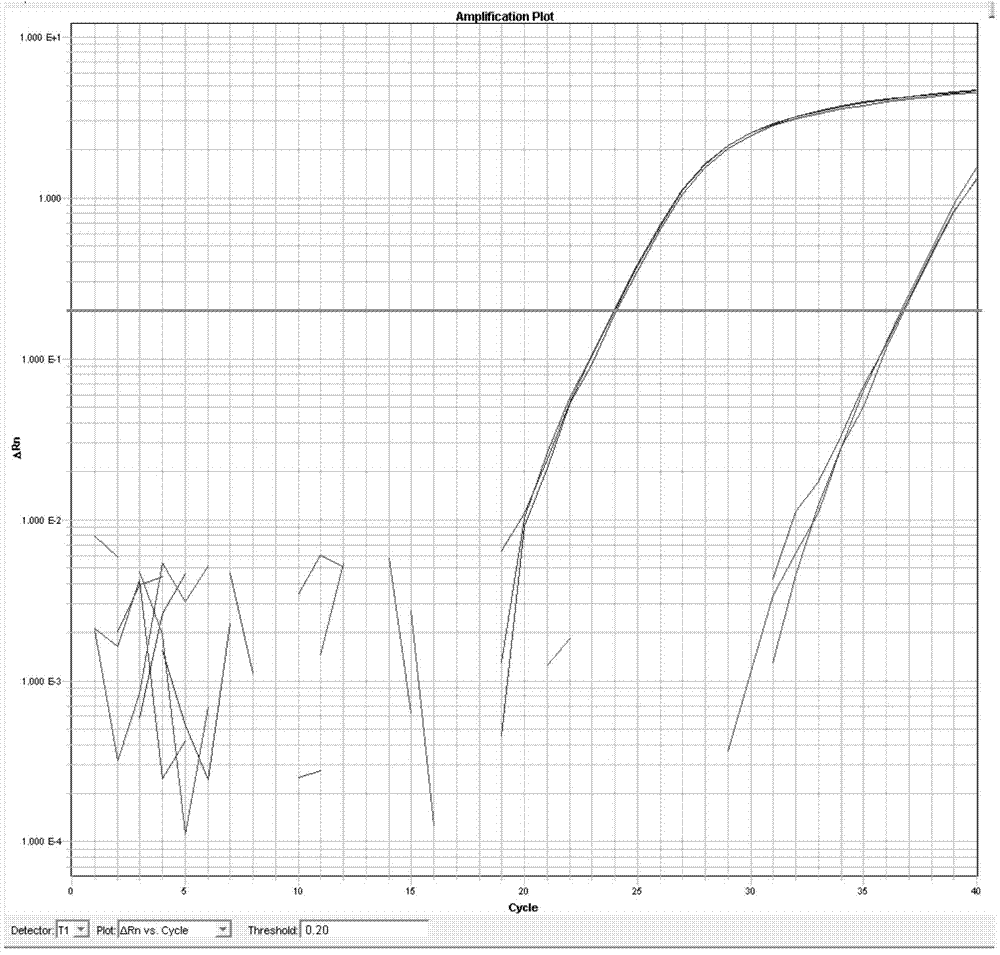
[0231] Using the probe library composed of SEQ ID NO. 1 to SEQ ID NO. 10 obtained in Example 1, the HCT-116 cell line (human colorectal cancer cell line, derived from ATCC cells) was detected by the same method as in Example 1. Library, catalog number: ATCC-CCL-247) KRAS gene, a single base mutation was found in the KRAS gene of HCT-116 cell line, which is 38G> A. See the result Figure 4 .
[0232] After DNA extraction, PCR amplification and Sanger sequencing, the above mutations have been verified.
Embodiment 3
[0233] Example 3: Use the method of the present invention to enrich and detect the EGFR gene in cells
[0234] This example detects the EGFR gene of the K-562 cell line.
[0235] 1. Prepare the DNA sample bank of the K-562 cell line to be tested
[0236] Except that the K-562 cell line (human chronic leukemia cell line, from the ATCC cell bank, catalog number: ATCC-CCL-243) is different, the experimental operation is the same as the first step of Example 1.
[0237] 2. Prepare DNA probe library for EGFR gene
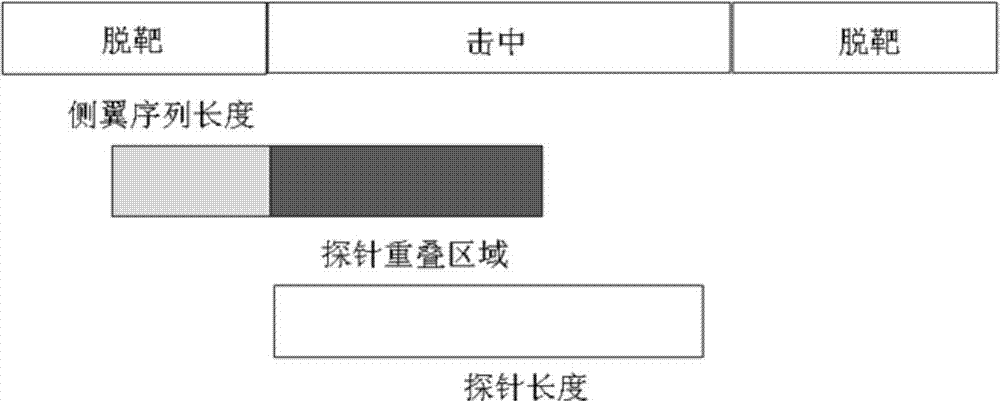
[0238] Except for the difference of EGFR mRNA (NCBI accession number: NM_005228.3), the experimental operation is the same as the second step of Example 1. Through the search of the human gene bank, it is ensured that the selected probe has a small off-target rate. After screening, first of all, 85 probes met the standard. Through IDT DNA Technologies, each probe was synthesized separately and analyzed by mass spectrometry to ensure the quality, and there was Biotin at the 5'en...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com