Method for constructing eukaryon gene knockout library by using CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9 system
A gene knockout and eukaryotic technology, applied in the fields of genomics and genetic engineering, can solve problems such as limited application range, system limitations, and phenotypic changes, and achieve cost reduction, low background value, and high positive rate Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
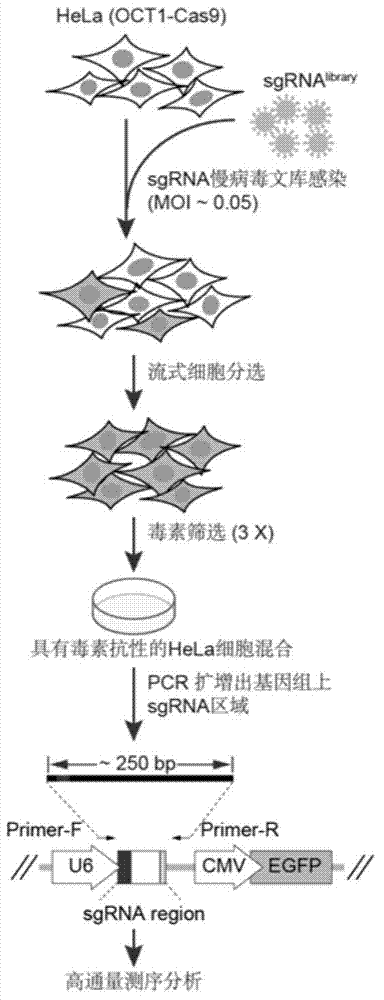
[0030] Example 1 Construction of CRISPER / Cas9 gene knockout library and screening of genes related to anthrax toxin cytotoxicity
[0031] 1. Design of library sgRNA
[0032] For 296 genes, 2-3 sgRNA target sites were found for each gene, see Table 1 for details.
[0033] Table 1 Gene composition of sgRNA library, sgRNA target region and primer sequence used to construct sgRNA plasmid
[0034]
[0035]
[0036]
[0037]
[0038]
[0039]
[0040]
[0041]
[0042]
[0043]
[0044]
[0045]
[0046]
[0047]
[0048]
[0049]
[0050]
[0051]
[0052]
[0053]
[0054]
[0055]
[0056]
[0057]
[0058]
[0059]
[0060]
[0061]
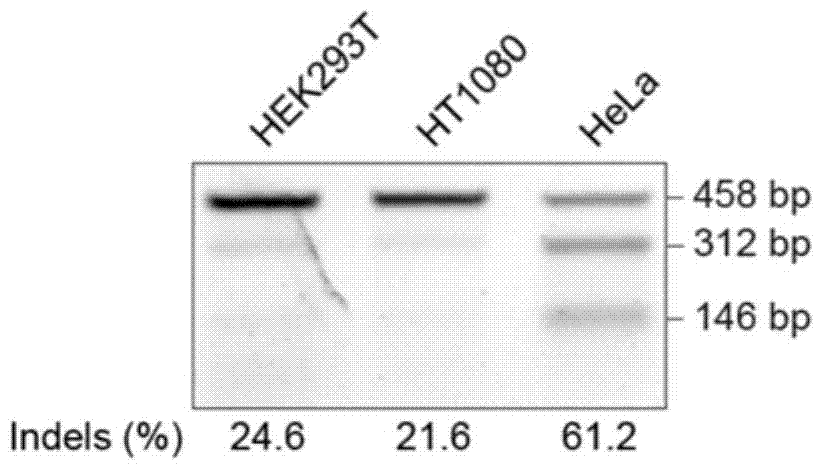
[0062] 2. Screening of HeLa cells highly expressing Cas9
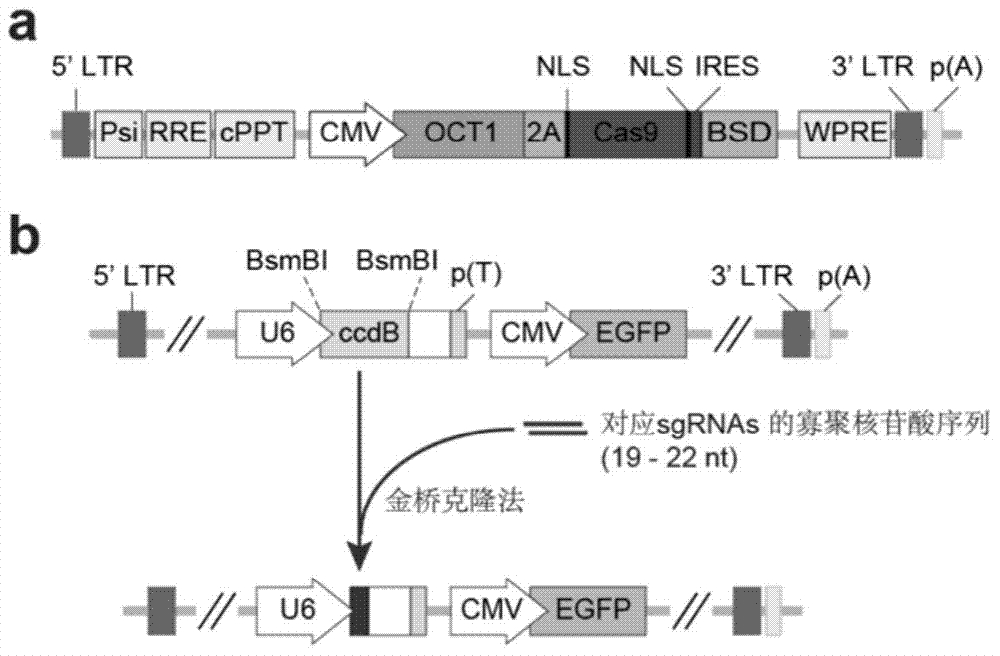
[0063] (1) The DNA sequences of OCT1 and Cas9 were connected by 2A and loaded onto the lentiviral vector pLenti-CMV-MCSSV-Bsd by the Gibson cloning method;
[0064] (2) Transfer the above-mentioned vectors i...
Embodiment 2
[0085] Example 2 Construction of CRISPER / Cas9 gene knockout library and screening of genes related to diphtheria toxin cytotoxicity
[0086] 1. Design of library sgRNA
[0087] For 296 genes, 2-3 sgRNA target sites were found for each gene, see Table 1 for details.
[0088] 2. Screening of HeLa cells highly expressing Cas9
[0089] (1) The DNA sequences of OCT1 and Cas9 were connected by 2A and loaded onto the lentiviral vector pLenti-CMV-MCSSV-Bsd by the Gibson cloning method;
[0090] (2) Transfer the above-mentioned vectors into the target cells by packaging lentivirus infection;
[0091] (3) Add blasticidin to the culture medium of the above cells for screening, and select a single clone stably expressing Cas9;
[0092] 3. Construction of Library Plasmids
[0093] (4) Look for the sgRNA action site, the sequence is 5'-G-Nx-NGG-3', where 19≤x≤22;
[0094] (5) Synthesize sgRNA monomers targeting the above-mentioned action sites. For each site, the sequence is, forward: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com