Dual-gene micro RNA fluorescent quantitative internal reference and application of primer in shellfish development samples
A fluorescent quantitative and dual-gene technology, applied in the field of molecular biology, can solve the problems of high cost and unsuitability for miRNA function analysis, and achieve the effect of low cost, high sensitivity and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
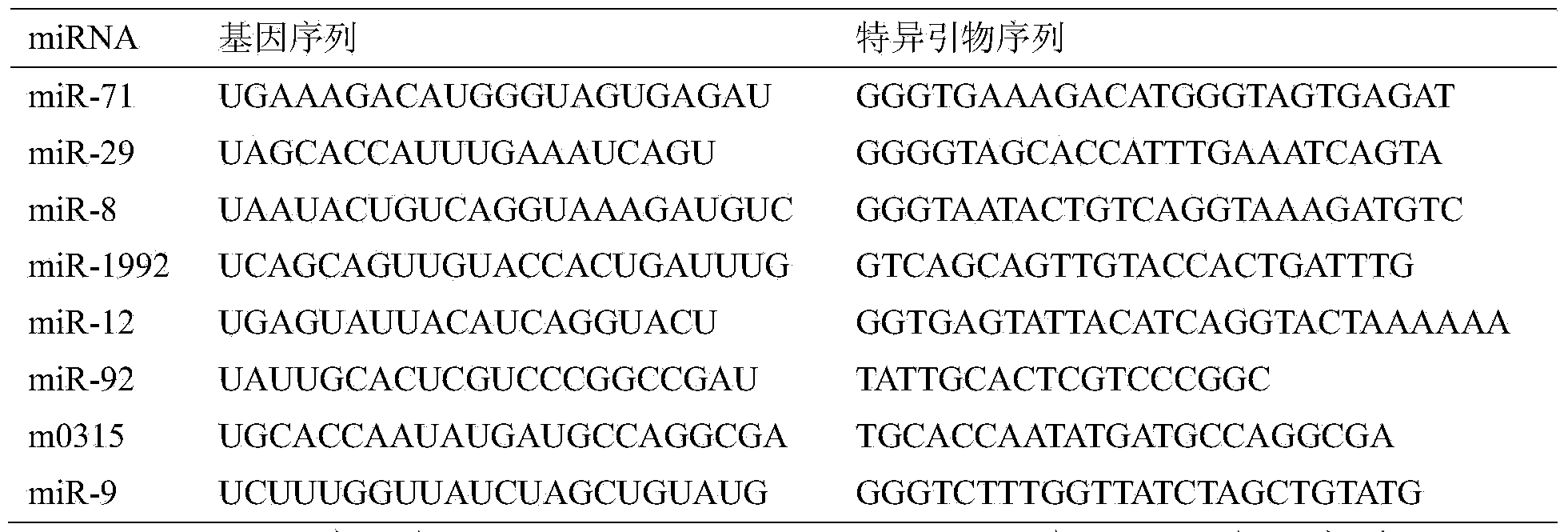
[0043] Selection of candidate internal reference genes: We performed microRNA sequencing on samples of Oyster oyster at different developmental stages, and obtained some newly identified microRNAs and their expression profiles. Based on the miRNA expression profiling data, we selected 6 microRNAs with relatively stable expression as candidate genes (ie, miR-8, miR-12, miR-29, miR-71, miR-92, miR-1992). At the same time, the commonly used microRNA internal reference gene 5.8S RNA, snRNA U6 and snRNA U1 were used as candidate genes.
[0044] The miRNA sequence (see Table 1) and sequencing expression profile of Oyster oyster are the results given by the RNA samples provided by us and sequenced and analyzed by Shenzhen BGI Institute. The following is a brief introduction to the method of deep sequencing analysis. For details, please refer to the literature [Huang J, Hao P, Chen H, Hu W, Yan Q, et al. (2009) Genome-wide identification of Schistosoma japonicum microRNAs using a deep...
Embodiment 2
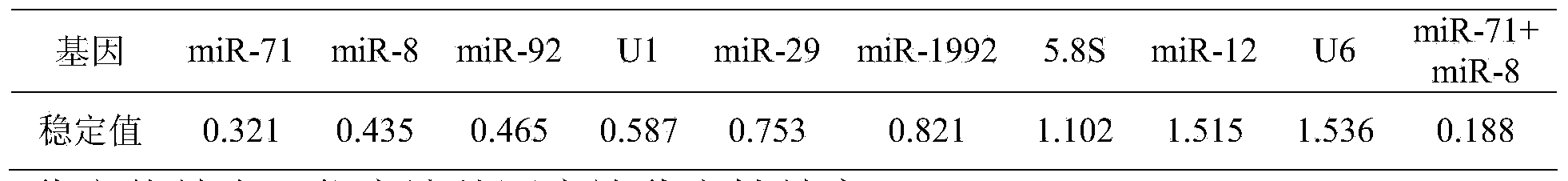
[0049] Evaluate the expression stability of candidate internal reference genes by fluorescent quantitative PCR: design specific primers according to the candidate gene sequence, perform fluorescent quantitative PCR in developmental samples, and obtain the expression data of each candidate gene at each developmental stage—Ct value (threshold cycle, The number of cycles experienced when the fluorescent signal in each reaction tube reaches the set threshold value). After converting the Ct value data with formula 2-Ct, use the most commonly used excel algorithm NormFinder for internal reference screening to calculate the stable value of each candidate gene. The smaller the stable value, the more stable the gene expression. For the specific algorithm and principle of NormFinder, please refer to the literature [ANDERSEN, C.L., JENSEN, J.L.&ORNTOFT, T.F.2004Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify ge...
Embodiment 3
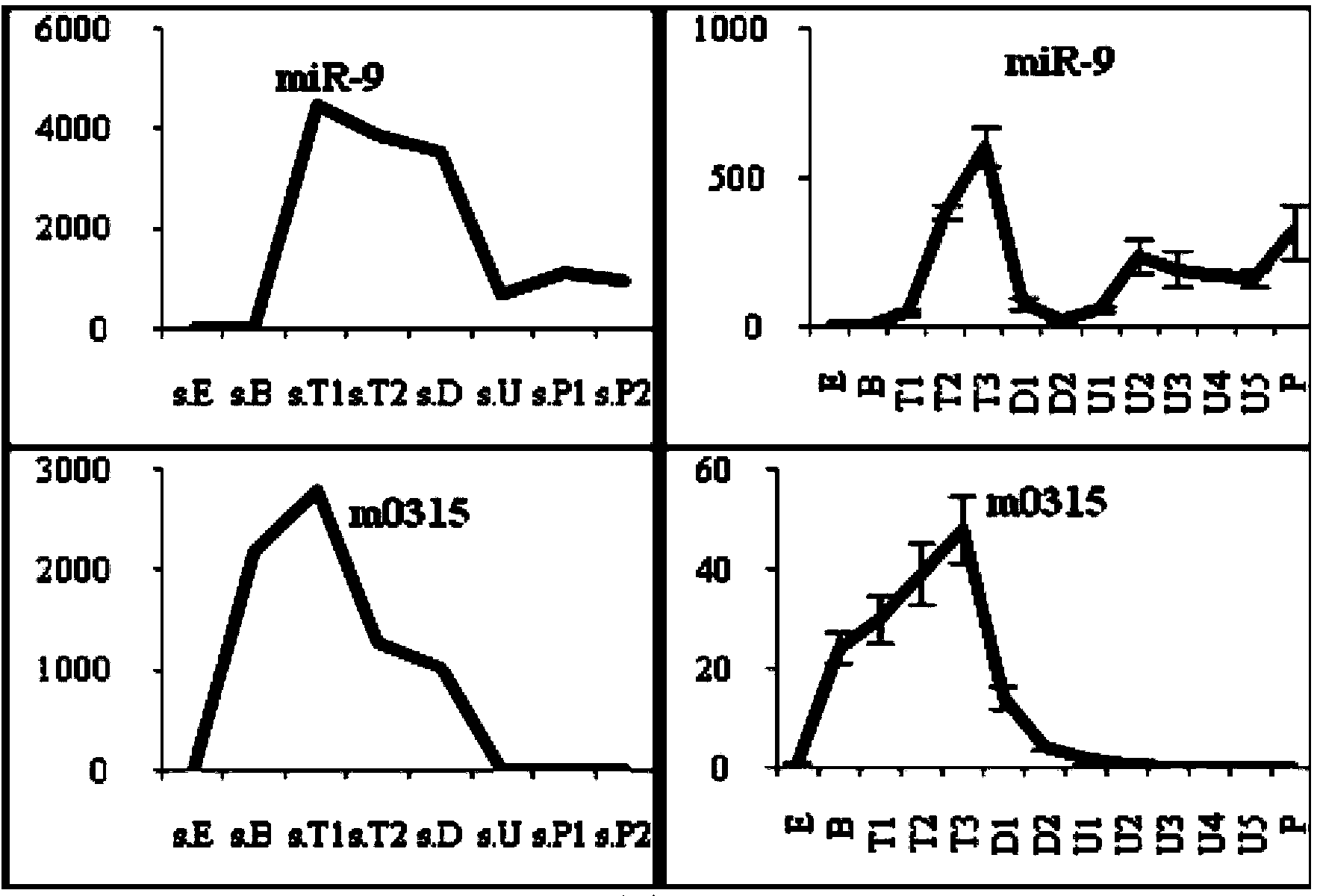
[0068] The two-gene combination is used as an internal reference, and the expression Ct value of the two-gene internal reference is the arithmetic mean of the expression Ct values of miR-71 and miR-8 genes in a certain developmental sample. Using the method based on SYBR Green dye fluorescence quantitative PCR, the relative quantitative analysis of two randomly selected long oyster miRNAs (miR-9 and m0315) in 13 developmental samples was carried out, see the literature: 【Shi R, Chiang VL(2005) Facile means for quantifying microRNA expression by real-time PCR.Biotechniques255039:519-525.]. It was found that the results of fluorescent quantitative PCR using the double gene as an internal reference were highly consistent with the results of the sequencing expression profile, which proved the reliability and stability of the dual gene internal reference.
[0069] Relative quantification of long oyster miRNA:
[0070]Obtaining of developmental samples, RNA extraction and synthes...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com