Electroluminescence logic gate adopting adenosine monophosphate and adenosine deaminase as excimers
A technology of adenosine monophosphate and adenosine deaminase, applied in the field of electroluminescent logic gates
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0011] Embodiment 1 Treatment of multi-walled carbon nanotubes and preparation of multi-walled carbon nanotubes modified electrodes
[0012] 2g of multi-walled carbon nanotubes was crushed in 36% HCl for 6 hours, washed with water and filtered to neutrality, and then dissolved in 2.2mol / L HNO 3 After the supernatant was crushed for 1 hour, it was cultured in an oven at 90°C for one night, then it was crushed for 1 hour at room temperature, and washed with water until neutral. Take 1 mg of the treated multi-walled carbon nanotubes and dissolve them in 100 mL of water containing 1 mg of DMF, and then ultra-pulverize for 1 h until a black suspension is formed.
[0013] After rinsing, the oxidized glassy carbon electrode was immersed in 10mmol / L ethylenediamine containing 5mmol / LEDC and 8mmol / LNHS and slightly shaken for 5h. After washing again, the electrode was transferred to the suspension of multi-walled carbon nanotubes containing 5mmol / L EDC and 8mmol / L NHS, and the multi-w...
Embodiment 2
[0014] Example 2 Immobilization of single-stranded DNA on the surface of multi-walled carbon nanotube modified electrode
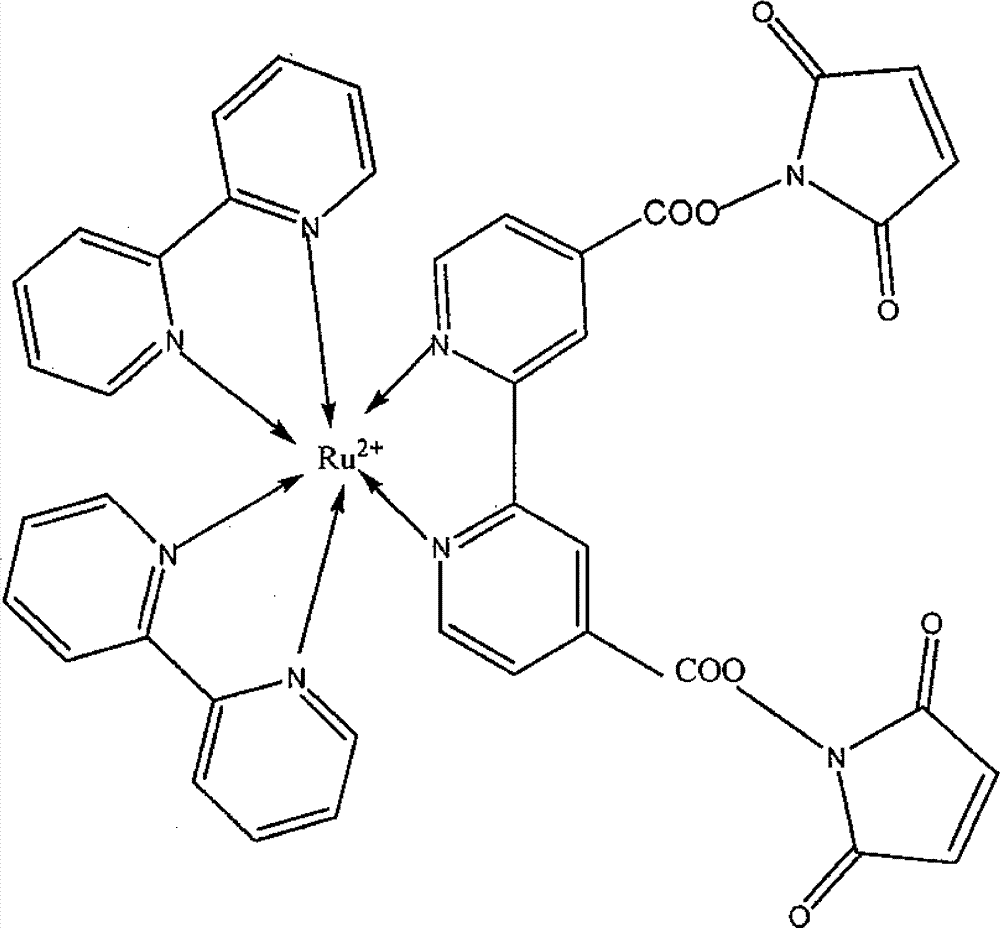
[0015] Take a certain amount of Ru(bpy) 2 (dcbpy)-S 1 ( figure 1 ) solution was mixed with 2.0mL0.1mol / L PBS buffer solution (pH7.0, 0.1mol / LNaCl), so that the concentration of ssDNA in the solution was 1×10 -6 mol / L, immerse the multi-walled carbon nanotube modified electrode in the above solution for 30 minutes with slight shaking. After fixing the electrode, use 0.2mol / L BR buffer solution (0.5% SDS, 0.04mol / LH 3 BO 3 , 0.04mol / L H 3 PO 4 , 0.04mol / L HAC, pH7.0) and secondary water were washed separately.
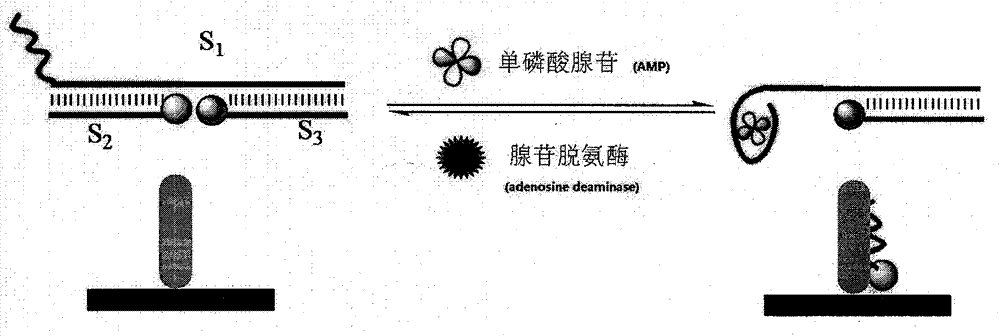
[0016] The improved operation takes a certain amount of Ru(bpy) 2 (dcbpy)-S 2 , S 1 and Fc-S 3 Mix in 2.0mL0.1mol / L PBS buffer solution (pH7.0, 0.1mol / L NaCl) so that the concentration of ssDNA in the solution is 1×10 -6 mol / L, after the hybridization is complete, add AMP to perform the same operation as above, which can make Ru(bpy) 2 (...
Embodiment 3
[0017] Example 3 Electrochemiluminescence detection
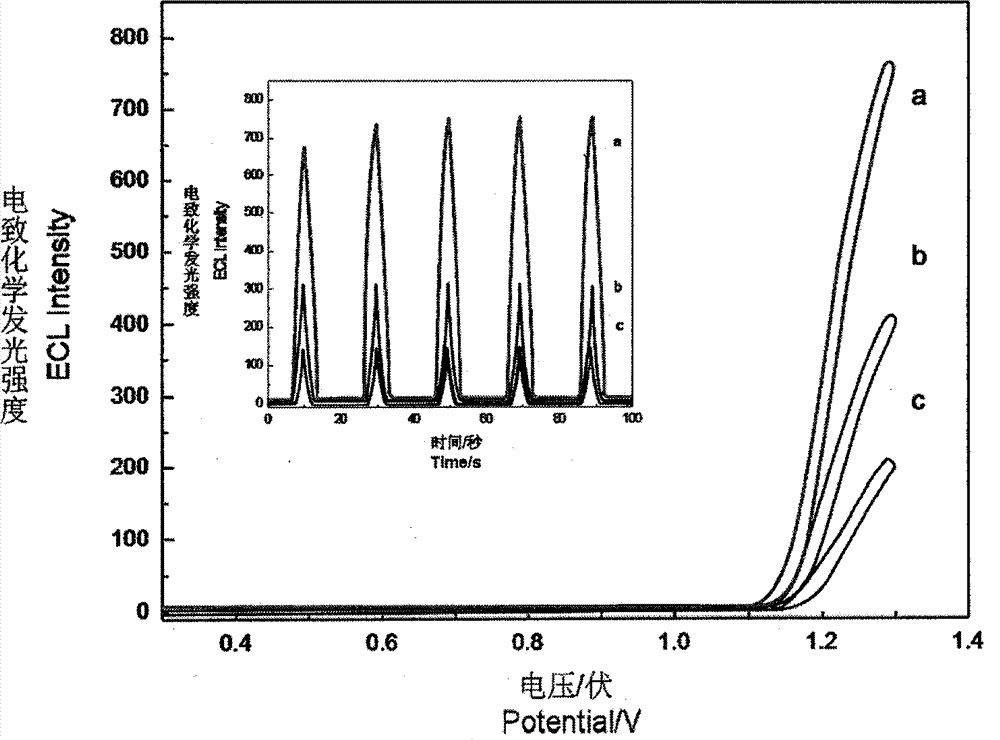
[0018] This experiment was carried out on the MPI-E electrochemiluminescence analysis system. Three-electrode system: with Ru(bpy) 2 (dcbpy)-ssDNA-modified multi-walled carbon nanotube modified electrode was used as the working electrode, a platinum wire electrode was used as the counter electrode, and Ag / AgCI (saturated KCI) was used as the reference electrode. The buffer solution is 2.0mL containing Ru(bpy) 2 (dcbpy)-ssDNA 0.1mol / LPBS buffer solution (pH7.0, 0.1mol / LNaCl), using cyclic voltammetry scanning (0.3-1.3V), the high voltage of the photomultiplier tube is 800V.
[0019] In the buffer solution, the concentration is 1×10 -6 mol / L of S 1 , S 2 and S 3 The reaction was stirred at a constant temperature of 37° C. for 30 minutes. 10 μL of 1 mmol / L AMP was added to the buffer solution, stirred and reacted at a constant temperature of 37° C. for 30 minutes, and then detected by electrochemiluminescence. After th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com