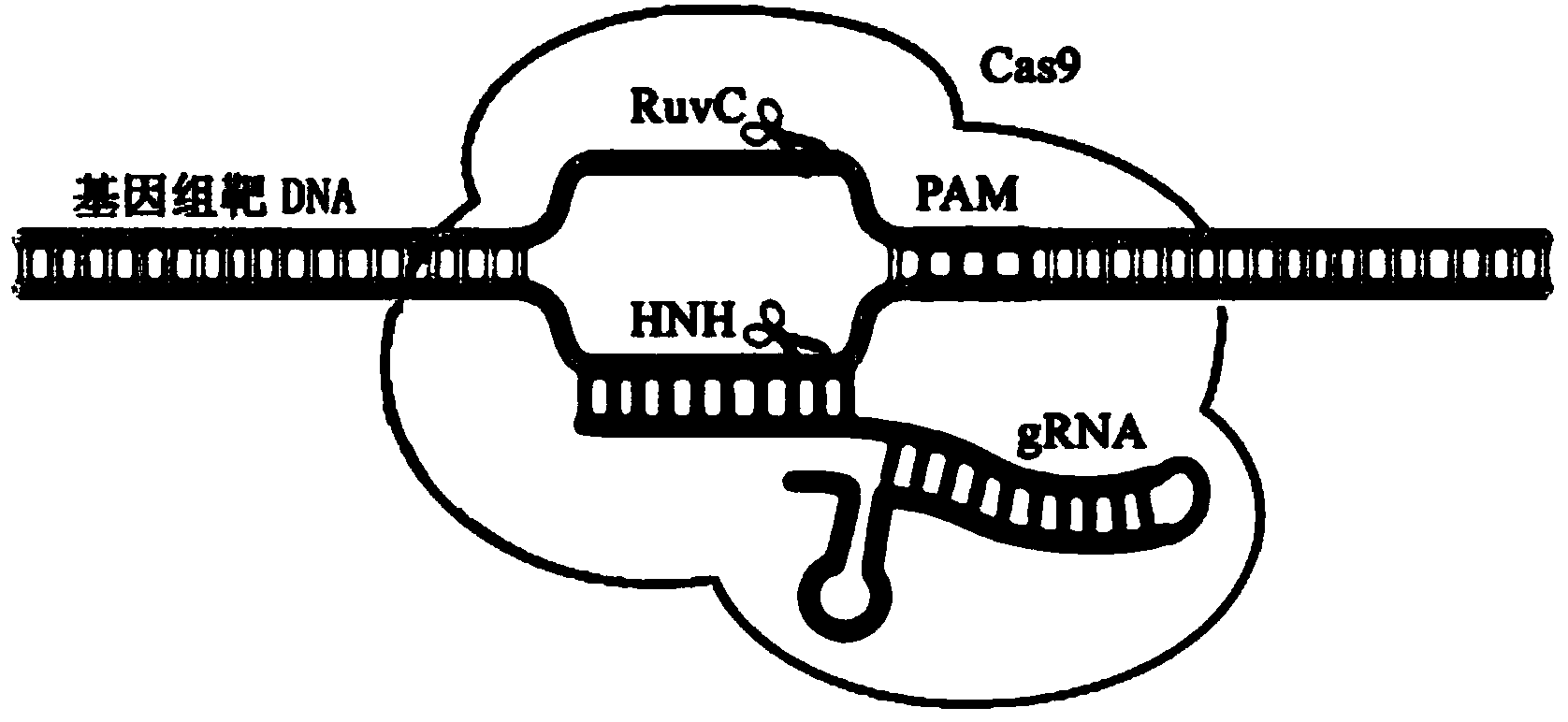
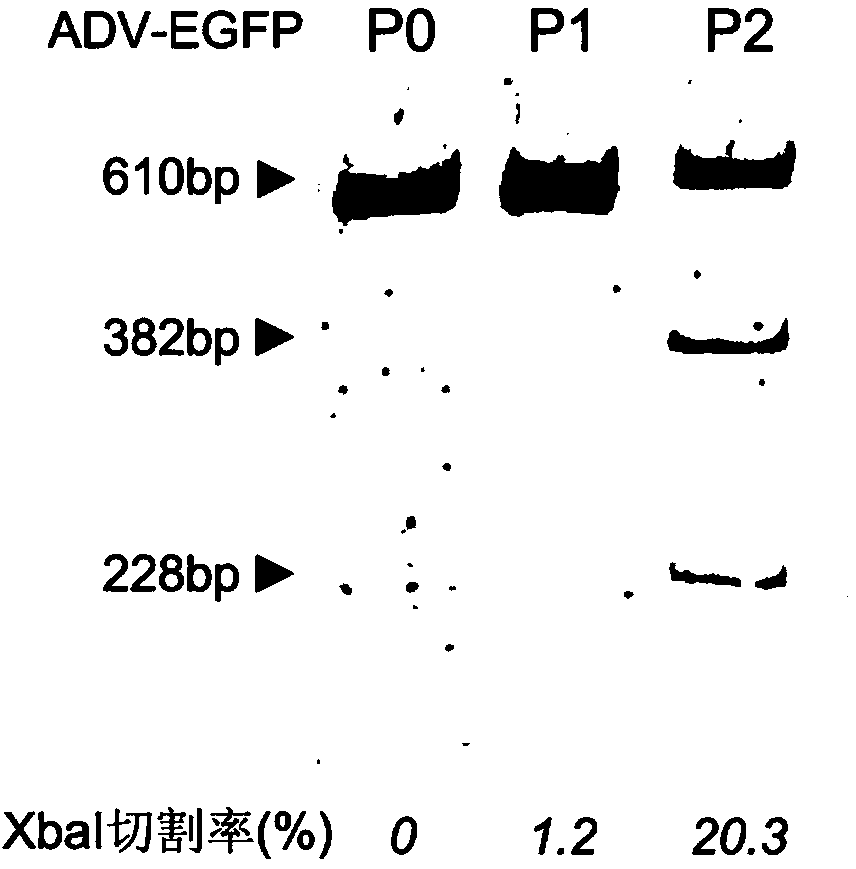
Site-specific modification and screening method for specific DNA (deoxyribonucleic acid) viral genome
A DNA virus and genome fixed-point technology, applied in DNA preparation, recombinant DNA technology, virus/bacteriophage, etc., can solve problems such as inability to meet precise insertion, mutation, and deletion of several bases
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] In this example, an adenovirus ADV-EGFP expressing jellyfish-enhanced green fluorescent protein was used, and the virus was constructed using the AdEasy system of Agilent Company. After the virus is amplified and purified, it is stored in sub-packages and stored in a deep-low temperature refrigerator for later use, avoiding repeated freezing and thawing. Take out a copy of the stored virus, and detect the virus ADV-EGFP titer by the plaque method, the titer is about 2x10 8 PFU / ml, in order to know the moi of infection (the number of viruses infected per cell - the multiplicity of infection).
[0061] A specific DNA virus genome site-directed transformation and screening method is achieved through the following steps:
[0062] Step 1, constructing a site-specific cleavage single-stranded nuclease system, specifically constructing a regularly repeating short palindromic sequence cluster-associated system
[0063](Clustered regularly interspaced short palindromic repeats...
Embodiment 2
[0079] The effect of the concentration of site-directed cutting single-strand nuclease system introduced into cells on the generation of insertion-deletion mutations
[0080] In this example, an adenovirus ADV-EGFP expressing jellyfish-enhanced green fluorescent protein was used, and the virus was constructed using the AdEasy system of Agilent Company. After the virus is amplified and purified, it is stored in sub-packages and stored in a deep-low temperature refrigerator for later use, avoiding repeated freezing and thawing. Take out a copy of the stored virus, and detect the virus ADV-EGFP titer by the plaque method, the titer is about 2x10 8 PFU / ml, in order to know the moi of infection (the number of viruses infected per cell - the multiplicity of infection).
[0081] Step 1, constructing a site-specific cleavage single-stranded nuclease system, specifically constructing a regularly repeating short palindromic sequence cluster-associated system
[0082] (Clustered regula...
Embodiment 3
[0087] Embodiment 3 comparative experiment
[0088] Wild-type Cas9 cleaves the double-stranded DNA of the viral genome through the following steps:
[0089] Step 1: After digesting and purifying the AG230 plasmid with BbsI, use T4DNA ligase (NEB) to connect to the DNA via P069 (5'-CAC CGC TGA AGC ACT GCA CGC CGT-3') and P070 (5'-AAA CAC GGC GTG CAG TGC TTC AGC-3') The double-stranded insert sequence formed by the annealing of two synthetic primers was used to construct the plasmid pCW175; the plasmid pCW175 was extracted by the endotoxin-free plasmid extraction kit for later use;
[0090] Step 2: Inoculate human embryonic kidney cells 293FT on 6-well cell culture plates at a seeding density of 6×10 5 After 24 hours, according to the instructions of liposome lipofectamine 2000 (Invitrogen), each well was transfected with 2 μg of plasmid AG230 or plasmid pCW175, and transfected for 24 hours to obtain transfected human embryonic kidney cells 293FT;
[0091] Step 3: After the tr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com