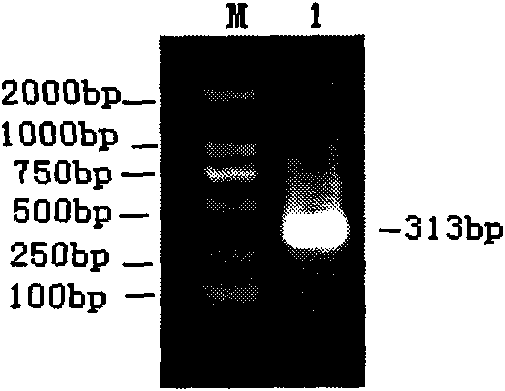
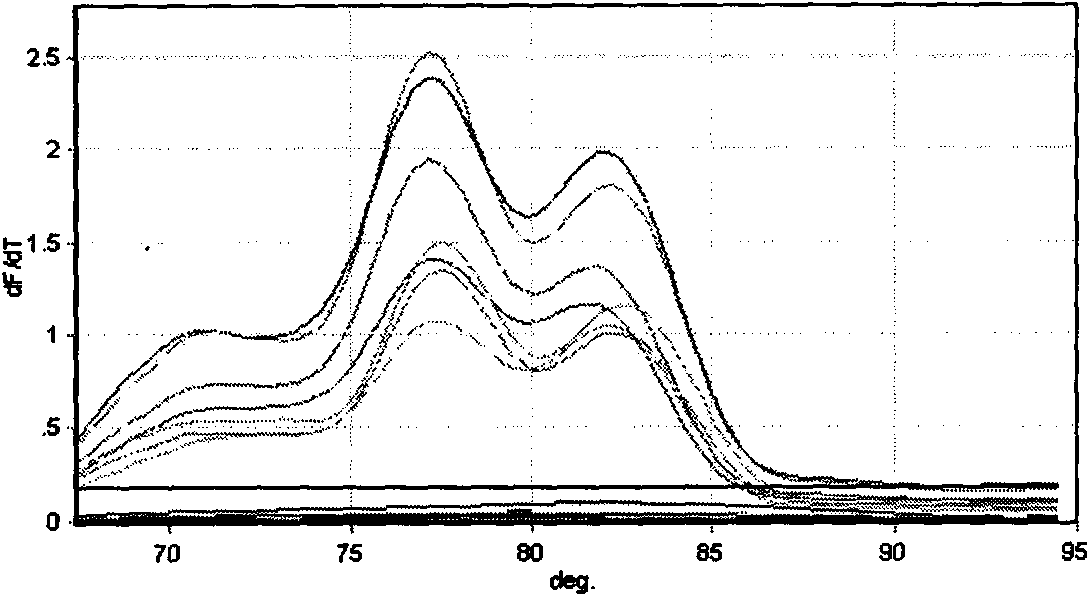
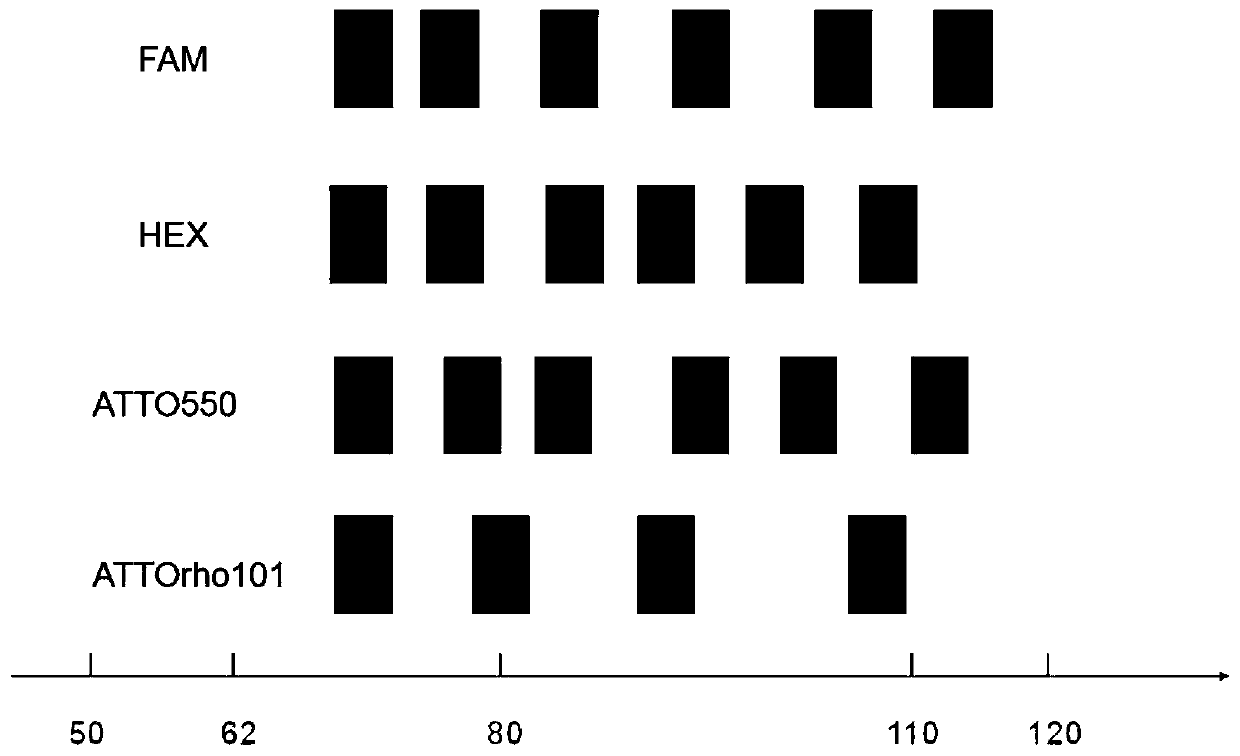
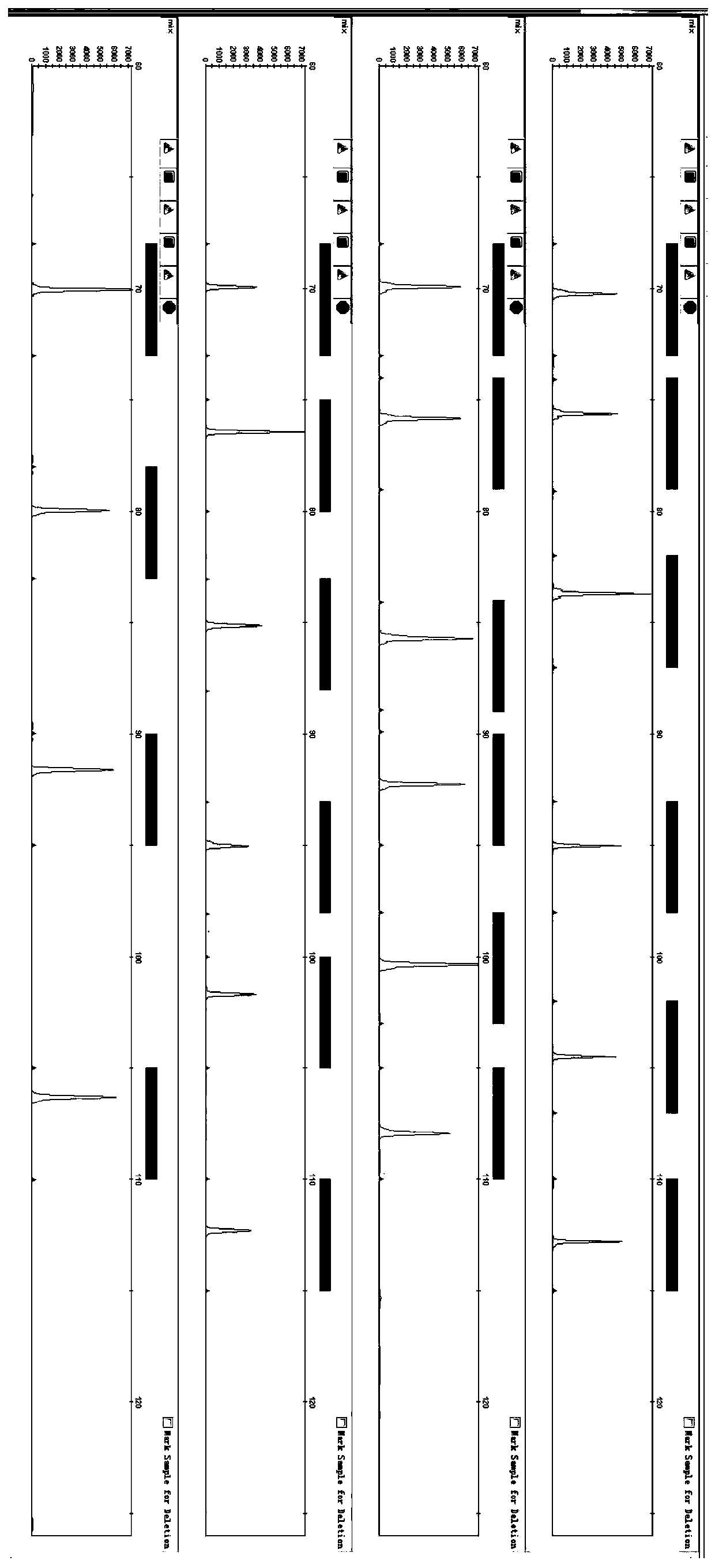
Patents
Literature
107 results about "Dna viral" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Treatment of neoplasms with viruses
InactiveUS20030044384A1Convenient treatmentReduce productionSsRNA viruses negative-senseBiocideDiseaseAnti viral response
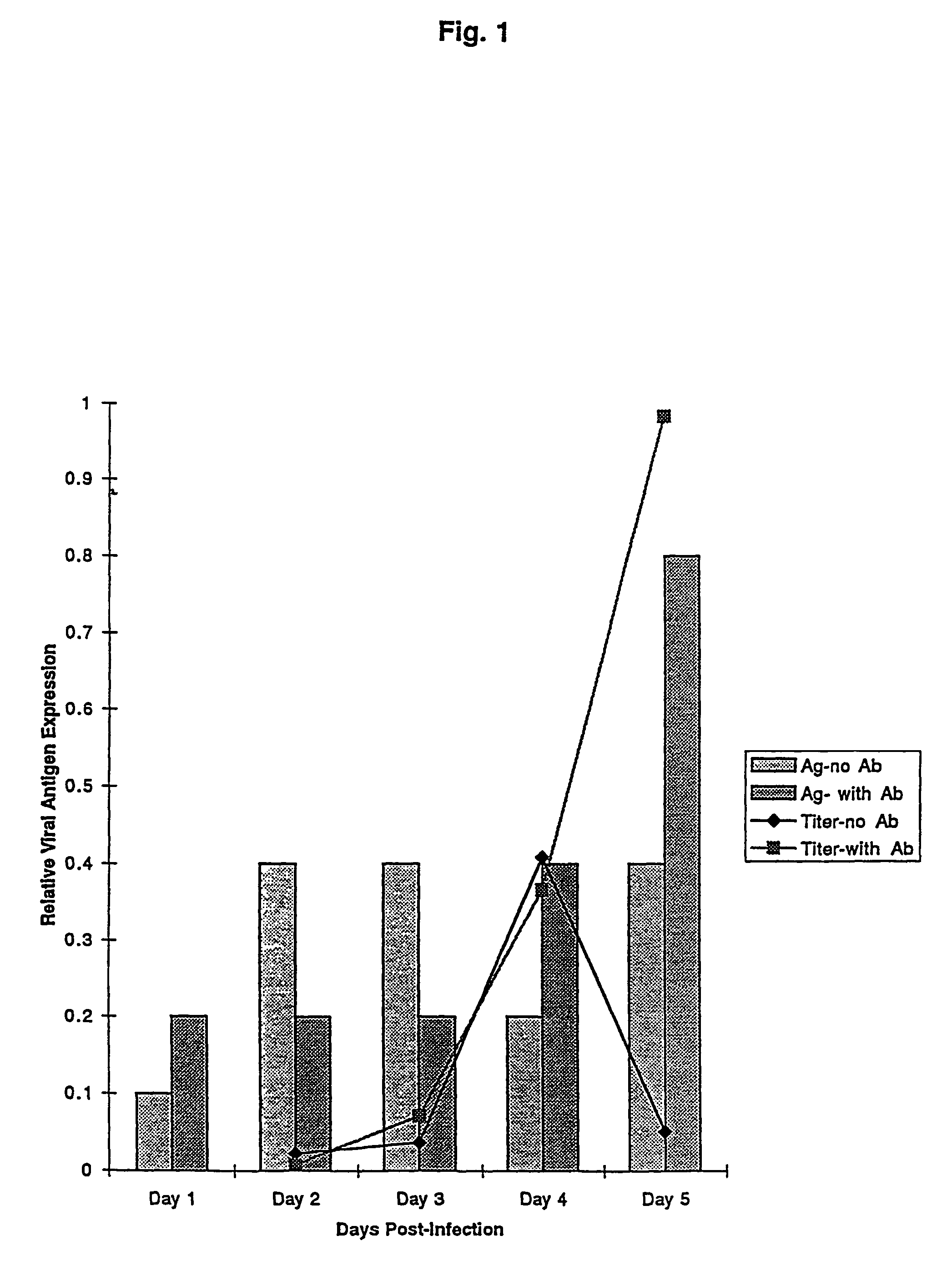
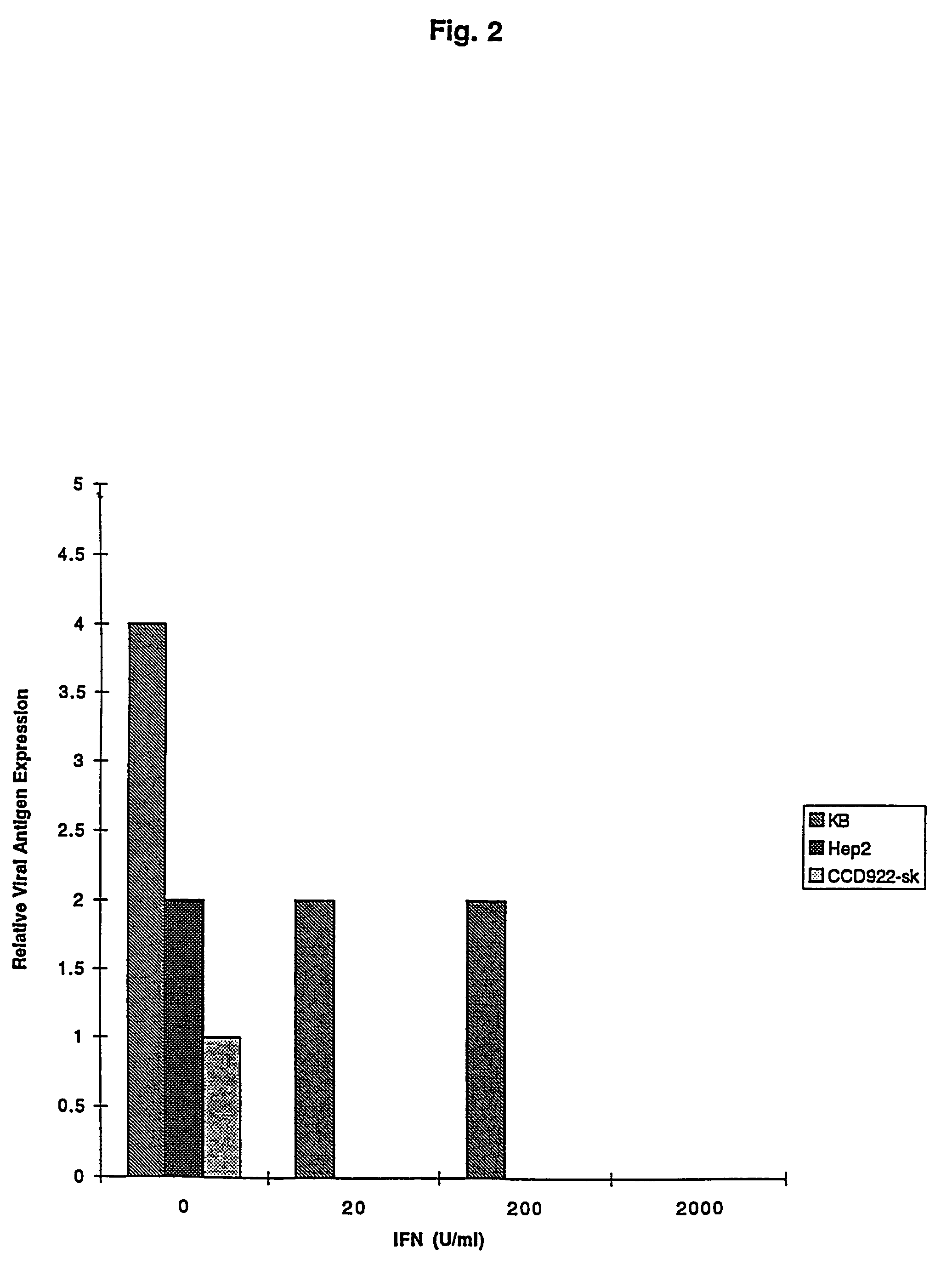
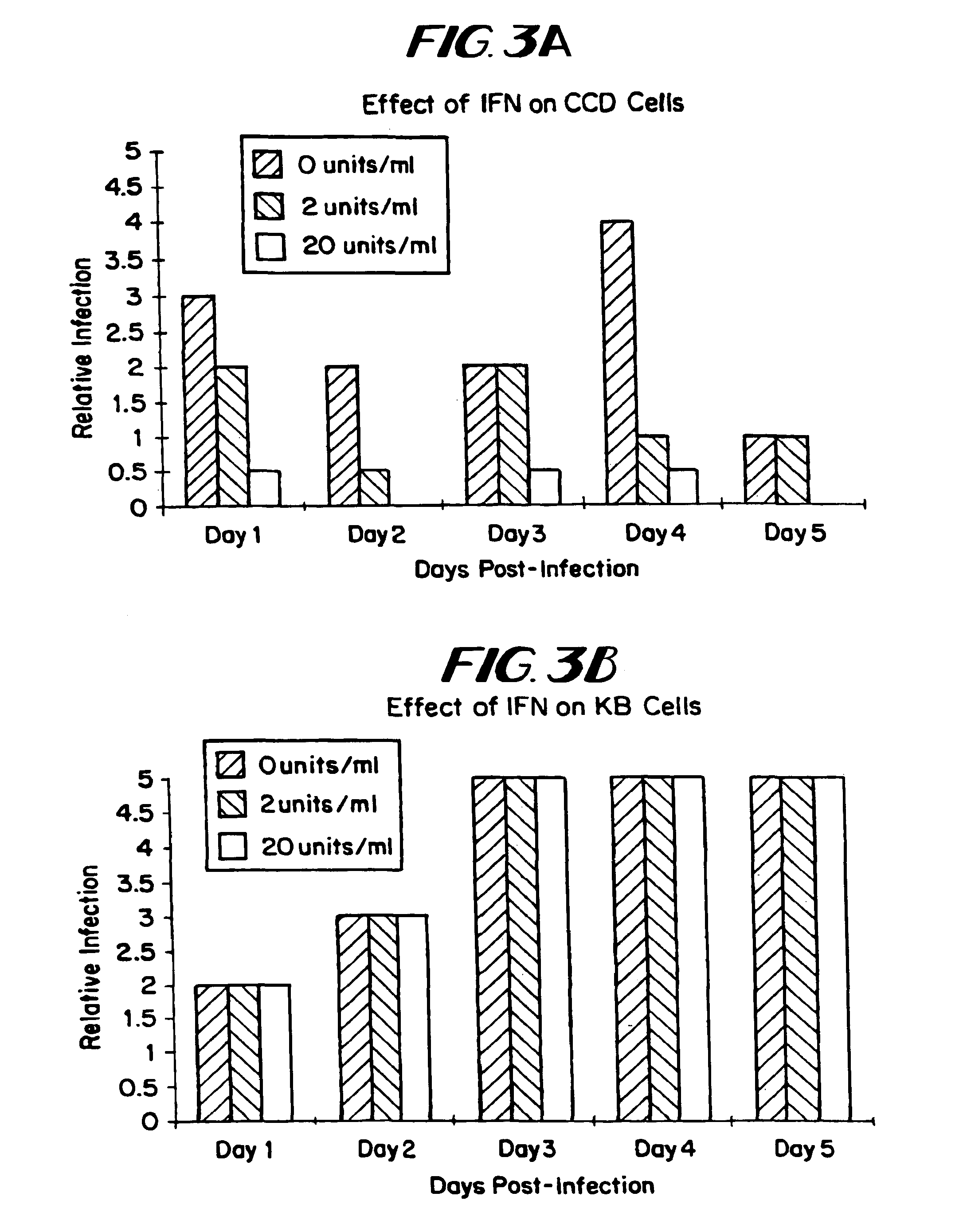
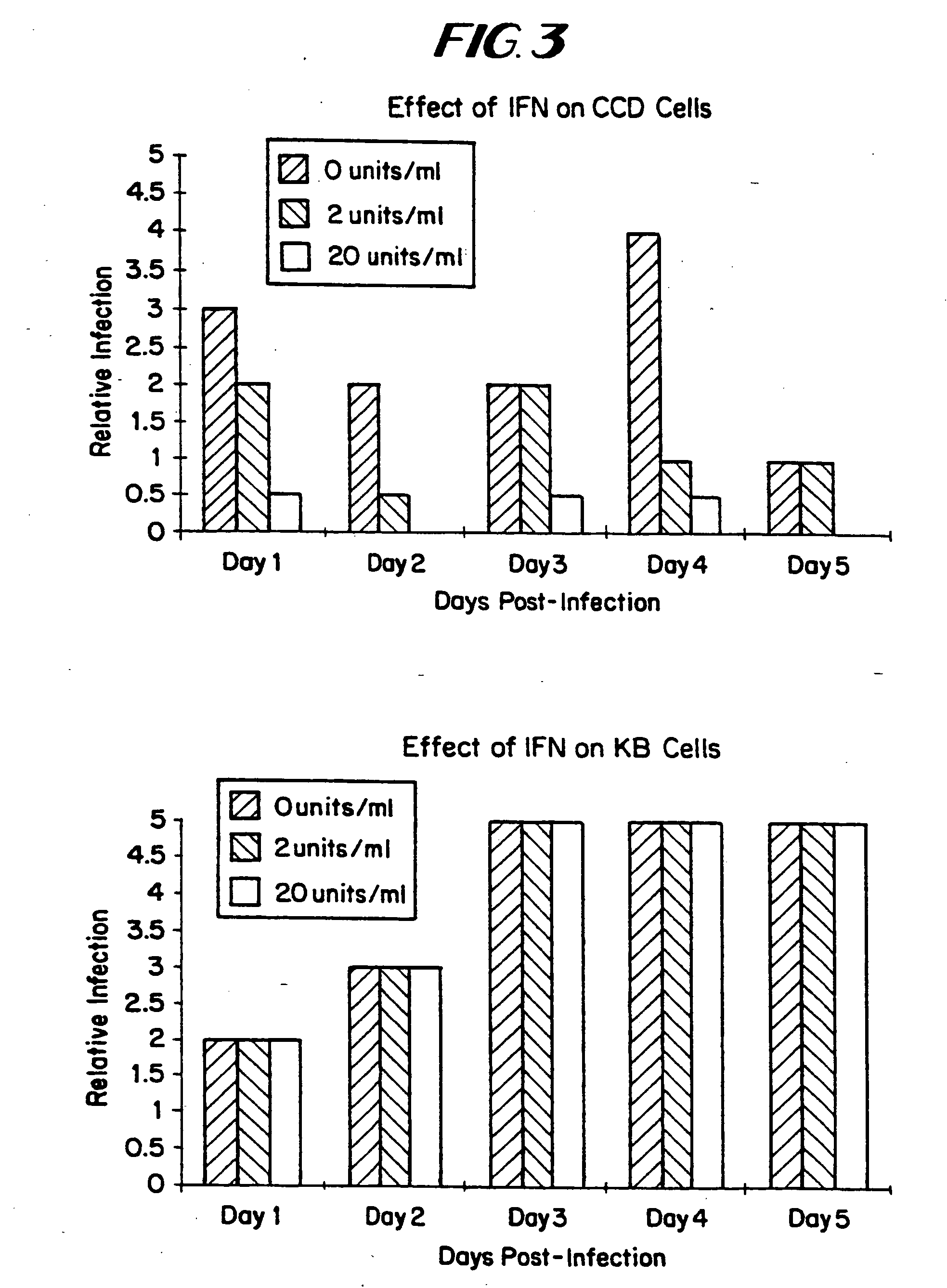
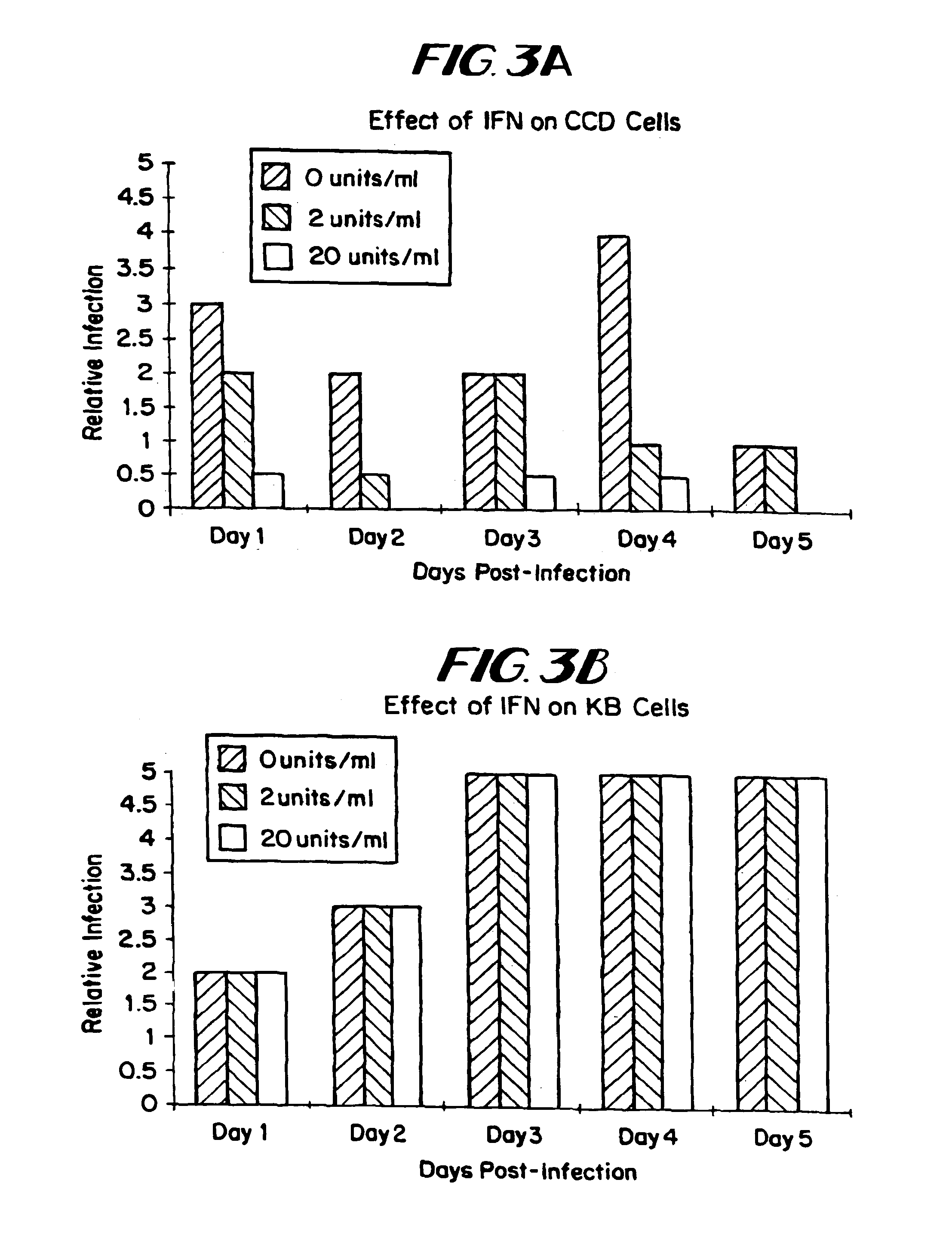
The subject invention relates to viruses that are able to replicate and thereby kill neoplastic cells with a deficiency in the IFN-mediated antiviral response, and their use in treating neoplastic disease including cancer and large tumors. RNA and DNA viruses are useful in this regard. The invention also relates to methods for the selection, design, purification and use of such viruses for cancer therapy.
Owner:PRO VIRUS
Retrovirus and viral vectors
InactiveUS6635472B1Prevent and attenuate diseaseEnhance immune responseGenetic material ingredientsVirus peptidesVirus-RetrovirusDna viral
This invention relates to the fields of genetic engineering, virus replication and gene transfer. More specifically, this invention relates to polynucleotide construct, recombinant virus, transposon, and their vectors, wherein an ori derived from a DNA virus capable of replicating in vertebrate cells is inserted into the retrovirus, allowing the retrovirus following the reverse transcription to efficiently replicate as extrachromosomal or episomal DNA without the necessity of integration into the host cell chromosome. Additionally, this invention relates to polynucleotide construct, recombinant virus, transposon, and their vectors replicating episomally without aid of an ori and related elements. Also, this invention encompasses preventive, therapeutic, and diagnostic applications employing said constructs, viruses and vectors.
Owner:RUBICON LABS
Treatment of neoplasms with viruses
InactiveUS20030165465A1Less weight lossConvenient treatmentSsRNA viruses negative-senseBiocideDiseaseAnti viral response
The subject invention relates to viruses that are able to replicate and thereby kill neoplastic cells with a deficiency in the IFN-mediated antiviral response, and their use in treating neoplastic disease including cancer and large tumors. RNA and DNA viruses are useful in this regard. The invention also relates to methods for the selection, design, purification and use of such viruses for cancer therapy.
Owner:WELLSTAT BIOLOGICS CORP
Lysine demethylase inhibitors for diseases and disorders associated with hepadnaviridae
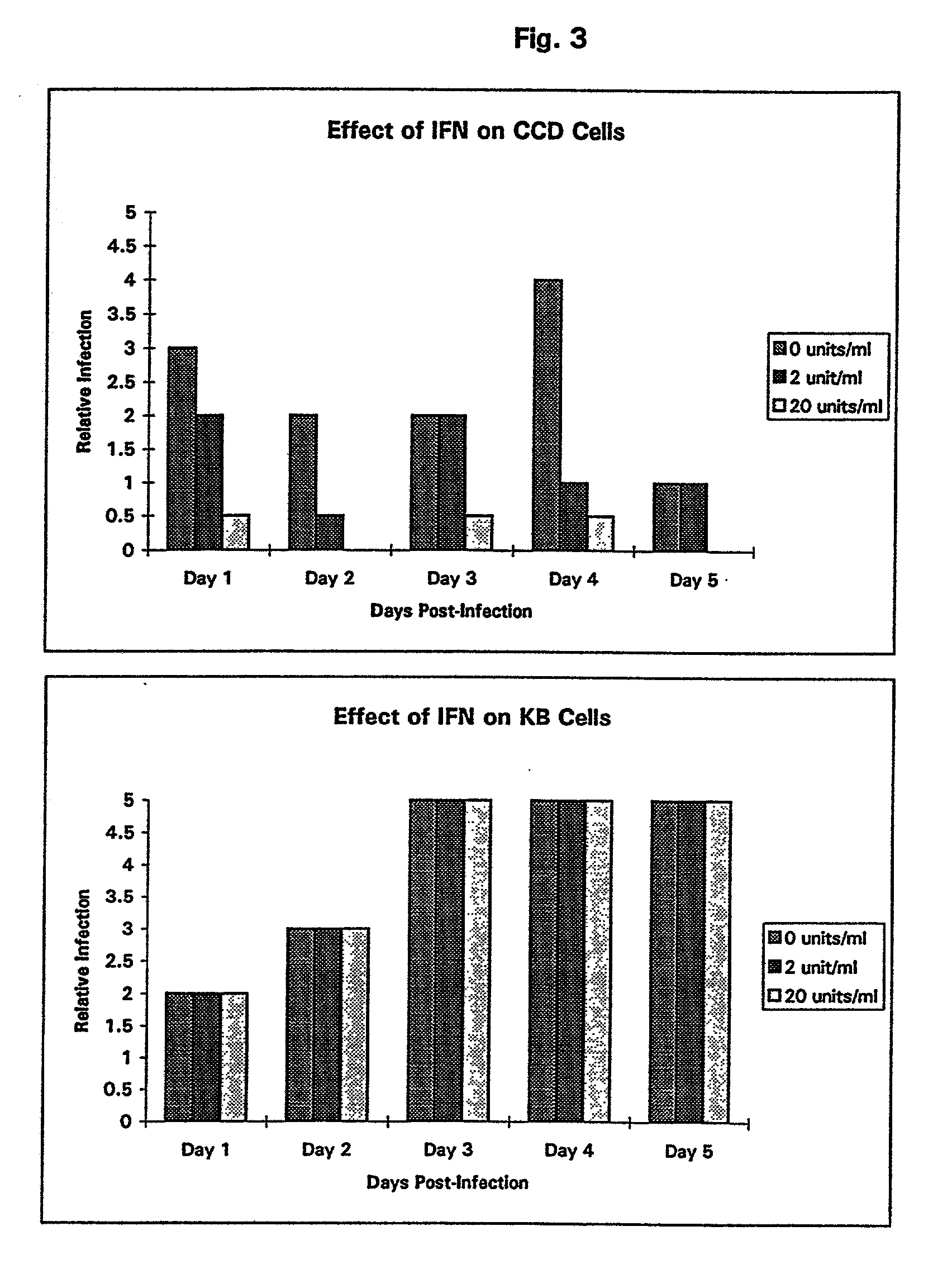
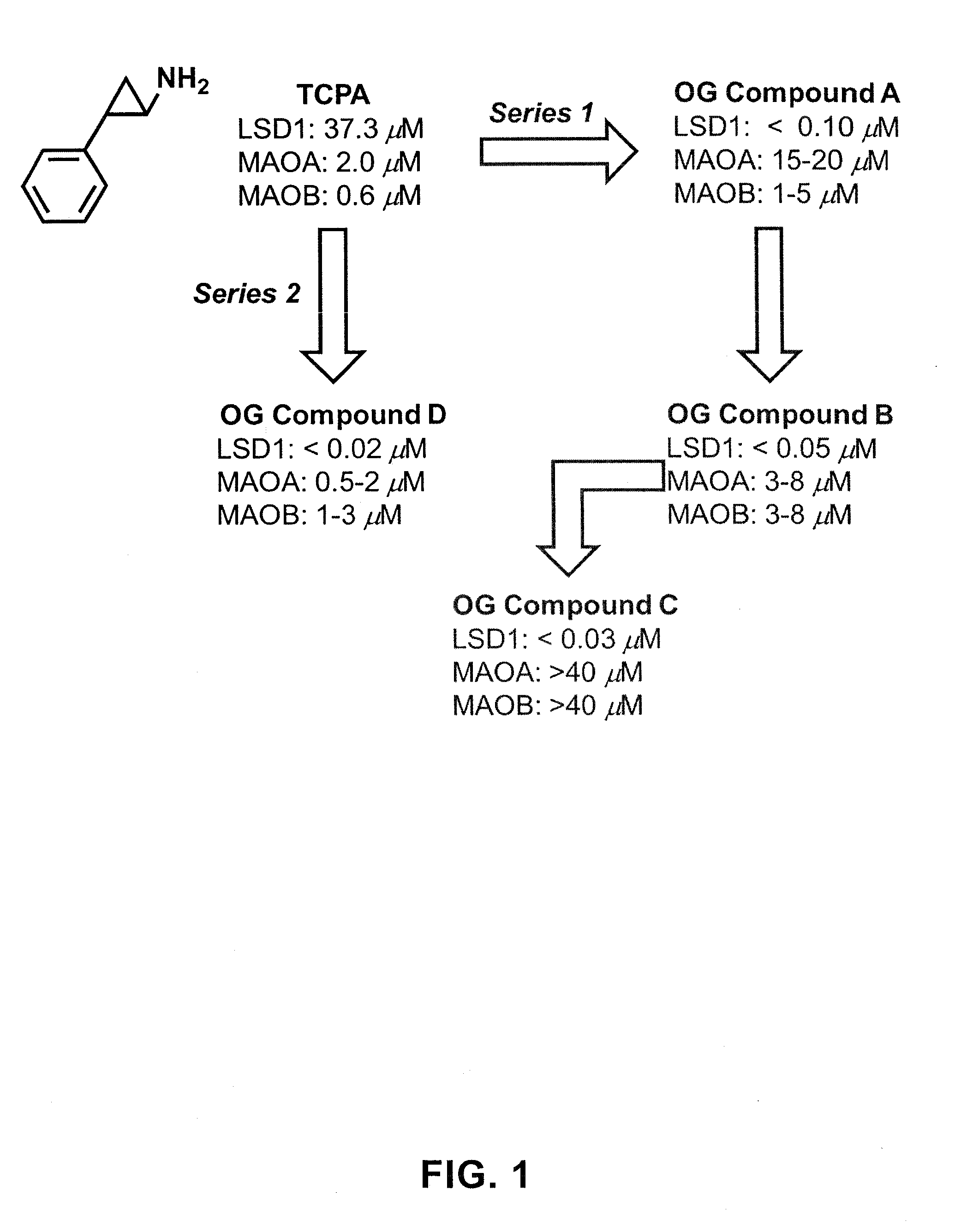
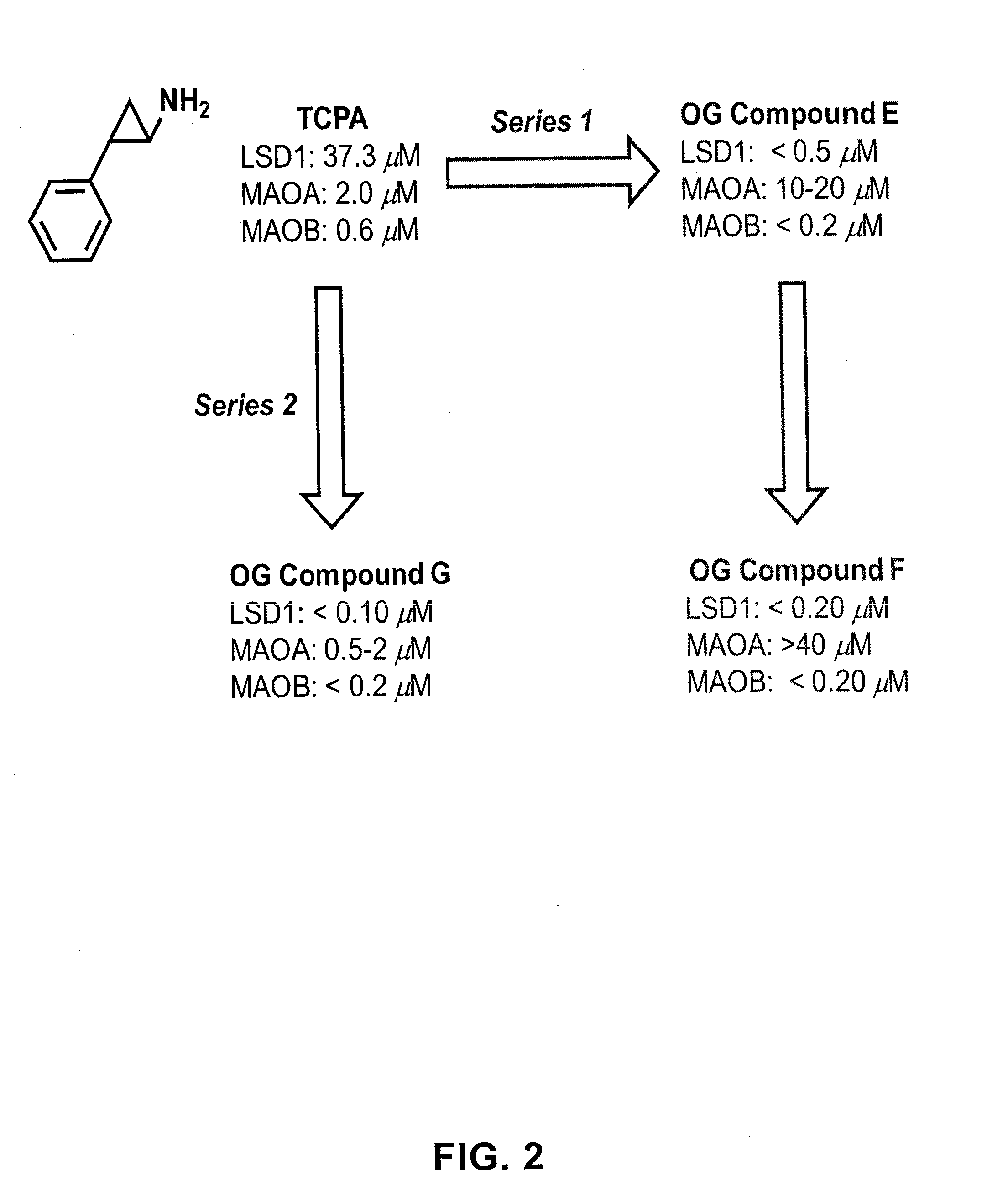
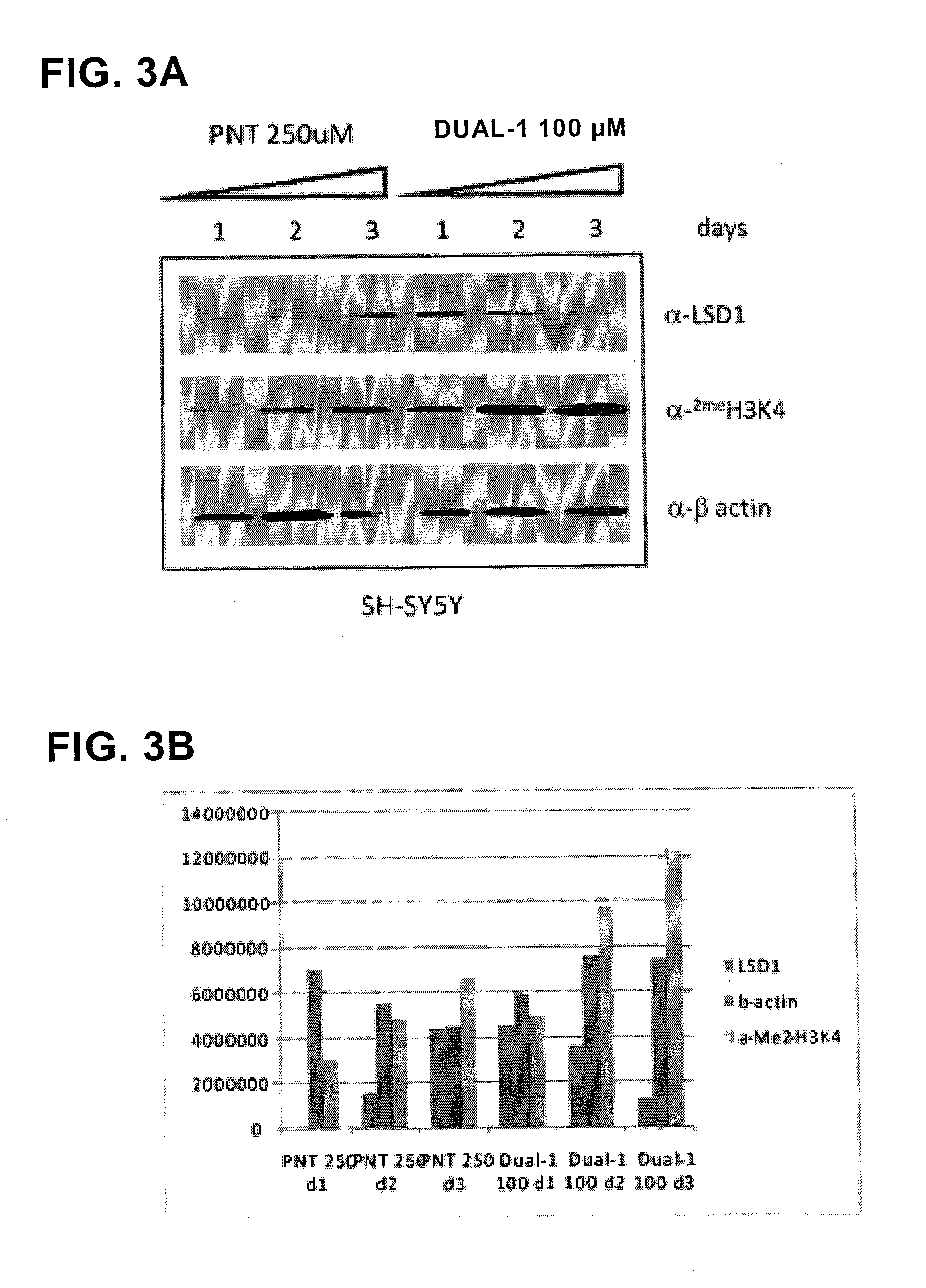
ActiveUS20130095067A1Symptoms improvedReduce probabilityBiocidePeptide/protein ingredientsMammalHepadnavirus
The present invention provides for the treatment or prevention of Hepadnaviridae infection and disease caused by Hepadnaviridae infection. In particular, the invention provides compositions and methods that affect the ability of Hepadnaviridae to utilize the host's cellular machinery as part of the virus' lifecycle. The invention relates to the discovery that interfering with the normal ability of viruses to utilize the host cell machinery with LSD1 inhibitors reduces HBV replication. Thus, the treatment and prevention of Hepadnaviridae infection and disease caused by Hepadnaviridae according to the invention comprises administering to an individual in need of treatment, a therapeutically effective amount of a LSD1 inhibitor. The individual in need of treatment can be a human or, e.g., another mammal.
Owner:ORYZON GENOMICS SA
Respiratory pathogen multi-detection reagent kit
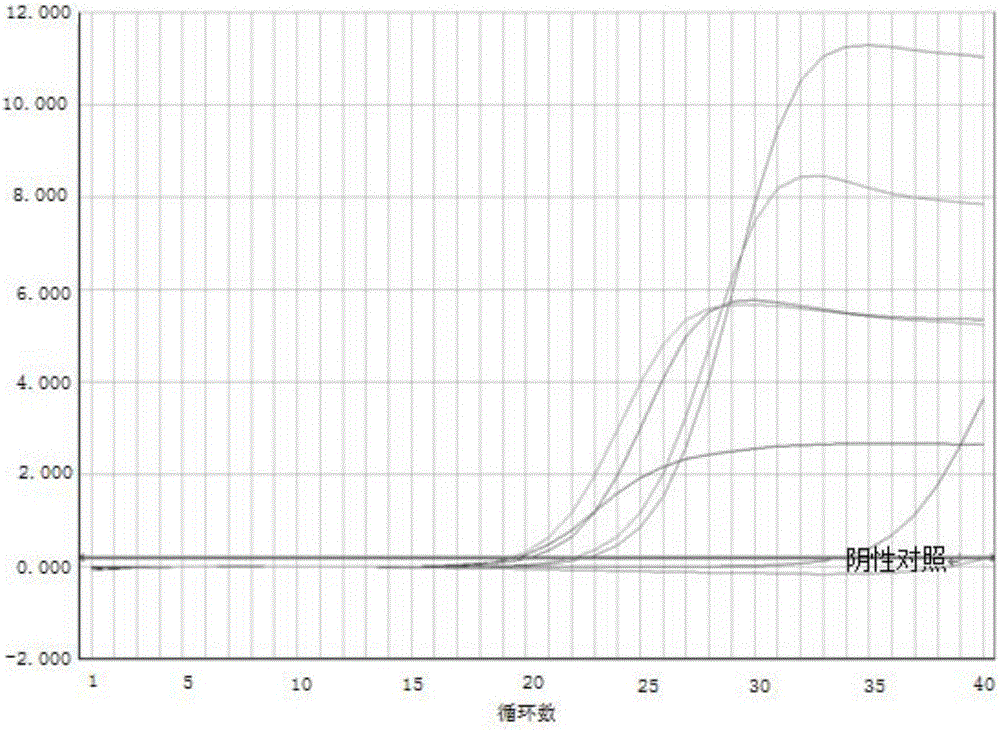
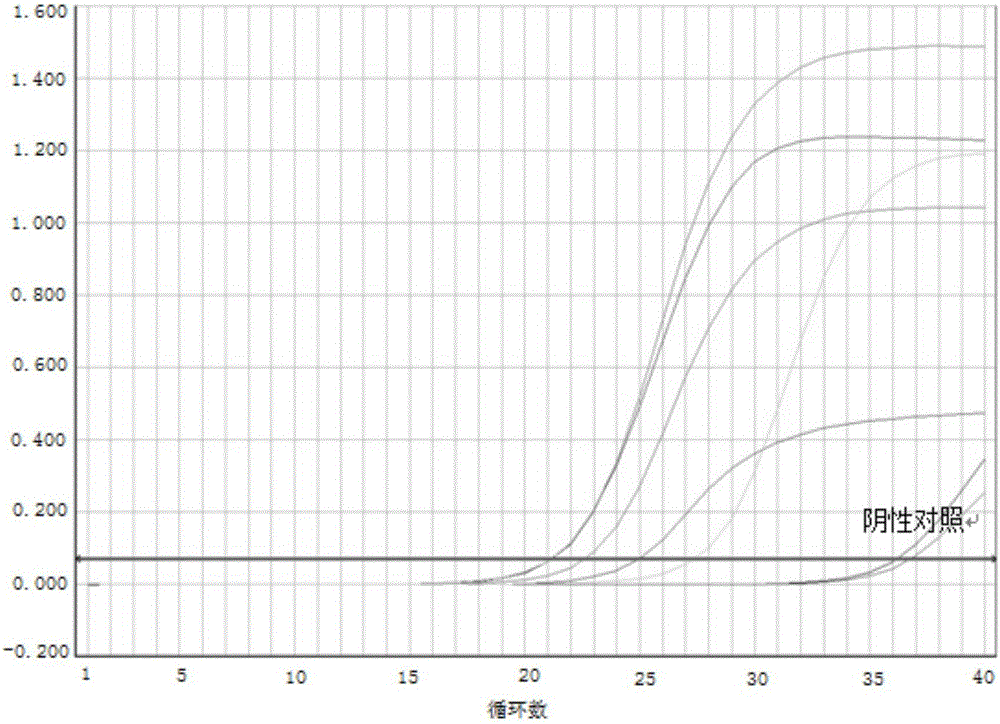
InactiveCN109355437AHigh detection sensitivityHigh sensitivityMicrobiological testing/measurementMicroorganism based processesCoronavirus 229EFluorescence
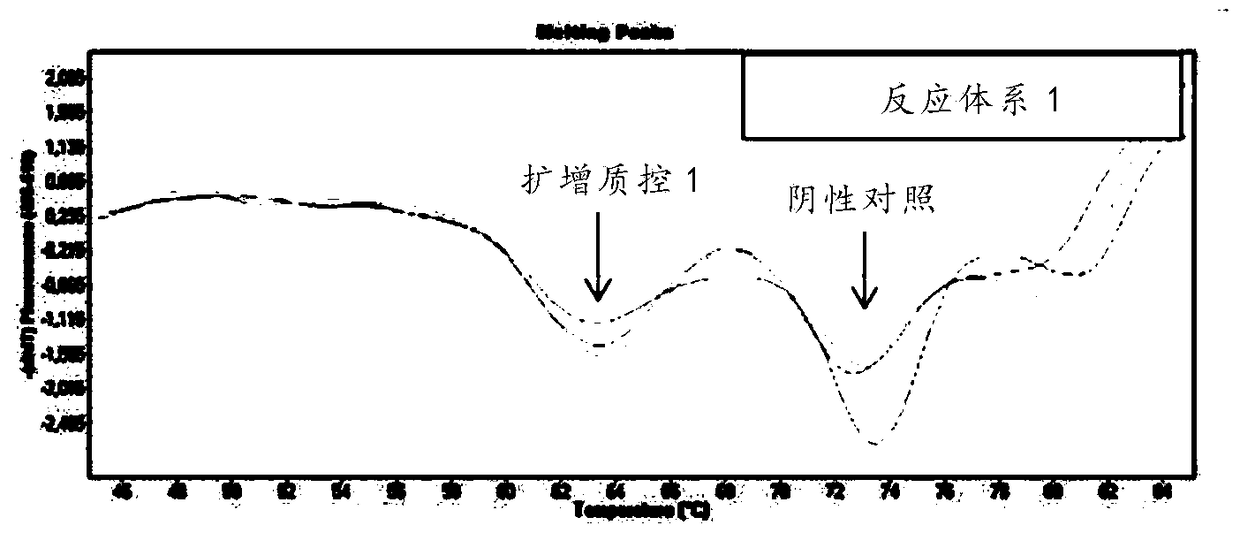
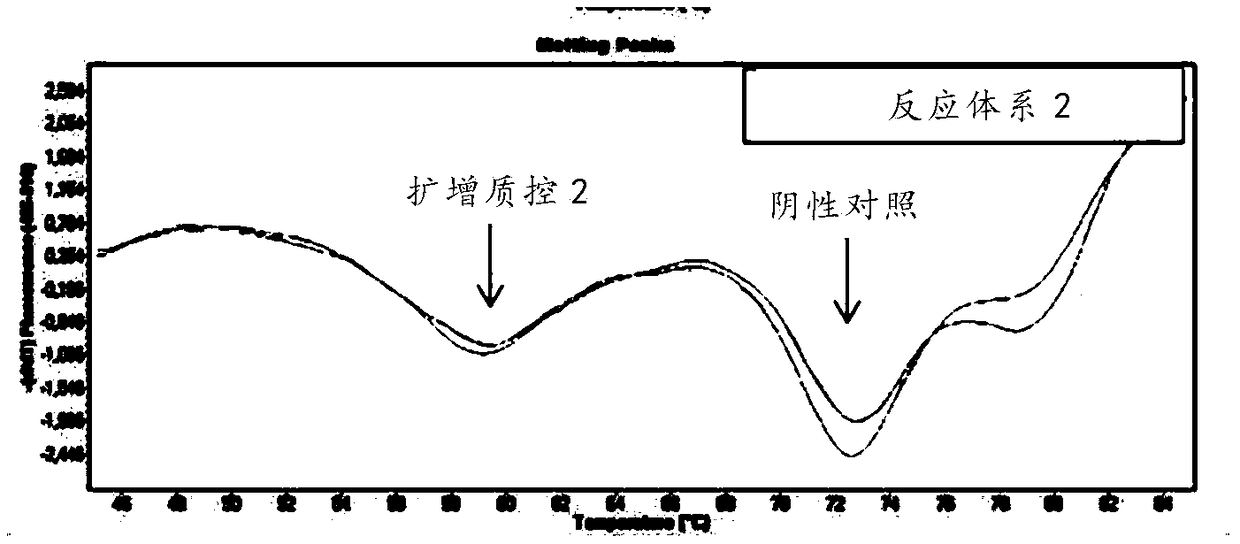
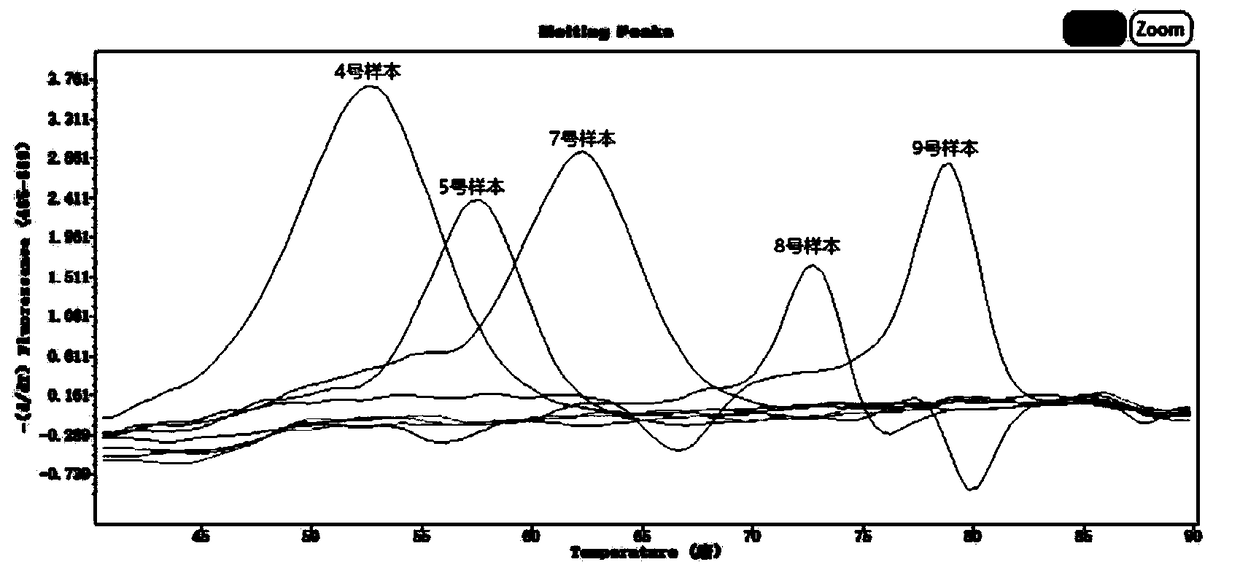
The invention discloses a respiratory pathogen multi-detection reagent kit. The respiratory pathogen multi-detection reagent kit has the advantages that the respiratory pathogen multi-detection reagent kit is based on multi-PCR (polymerase chain reaction) technologies, detection results can be determined by the aid of fluorescence resonance energy transfer via the melting temperature ranges, the respiratory pathogen multi-detection reagent kit can be used for qualitatively simultaneously detecting 16 types of respiratory pathogens, the 16 types of respiratory pathogens include 12 types of RNA(ribonucleic acid) viruses (influenza A viruses, influenza B viruses, H1N1 influenza A viruses, type A and type B respiratory syncytial viruses, type -1 / -2 / -3 parainfluenza viruses, type OC43 coronaviruses, type 229E coronaviruses, rhinoviruses and human metapneumovirus), 2 types of DNA (deoxyribonucleic acid) viruses (adenoviruses and bocavirus) and 2 types of bacteria (mycoplasma pneumoniae andbordetella pertussis), the respiratory pathogen multi-detection reagent kit is high in detection sensitivity, and the sensitivity even can reach 1 copy / reaction; the multi-detection reagent kit is good in specificity, and negative results of pathogens which have identical sampling sites and similar pathogenic mechanisms and are not in the detection range of the respiratory pathogen multi-detectionreagent kit can be obtained; the respiratory pathogen multi-detection reagent kit is short in operation time and easy to operate and can be used for quickly detecting the 16 types of respiratory pathogens in a single tube of a reaction system, the results are clear and are easy to interpret, and the like.
Owner:上海捷诺生物科技股份有限公司
Treatment of neoplasms with viruses
Owner:WELLSTAT BIOLOGICS CORP
Site-directed modification method for DNA viral genome
ActiveCN103397018ARealize fixed-point transformationQuick insertRecombinant DNA-technologyFermentationFreeze thawingRecombinant virus
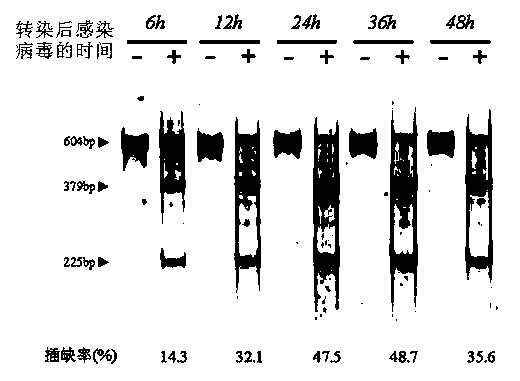
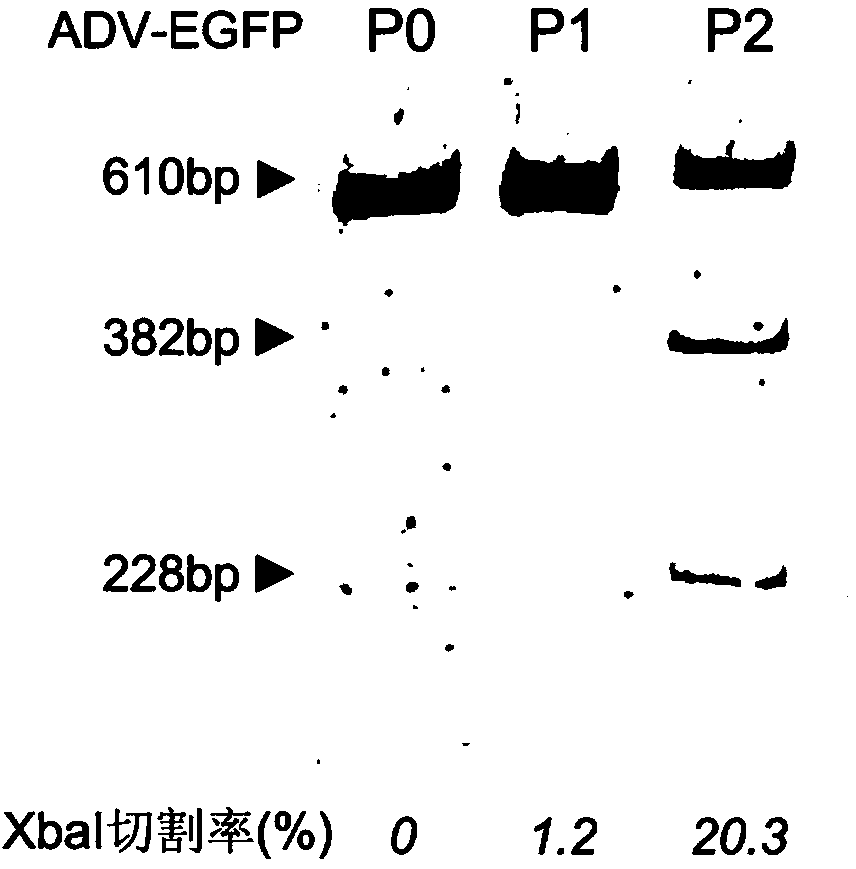
The invention provides a site-directed modification method for DNA viral genome, and the problems in the prior art are solved that induction of site-directed mutagenesis of DNA viral genome is difficult, the operation of inserting an exogenous fragment is complex, and recombination rate is lower. The site-directed modification method comprises: transfecting cells by a plasmid carrying a nuclease system, infecting by a virus, after the cells show pathological changes, collecting the cells with pathological changes, performing freeze-thaw or ultrasonic processing, and centrifuging, separating the liquid supernatant to obtain a progeny virus. The site-directed modification method is capable of realizing applications to screening of virus attenuated vaccine strains, construction of viral genetic carriers and an oncolytic virus, research on virus function sequences, and the like; during modification of the viral genome, the method helps to improve mutagenesis efficiency, accurately control DNA virus for genome site-directed mutagenesis and specific gene knockout, simplify operation steps of inserting the DNA virus carrier by an exogenous gene, and improve efficiency that the exogenous gene is integrated to the viral genome, so that the work of screening high-flux recombination viruses is convenient to conduct.
Owner:INST OF MEDICAL BIOLOGY CHINESE ACAD OF MEDICAL SCI
Compositions for the inactivation of virus replication and methods of making and using the same
ActiveUS20170049909A1Improve the level ofAccurate separationPolypeptide with localisation/targeting motifHydrolasesEpitopeNucleotide
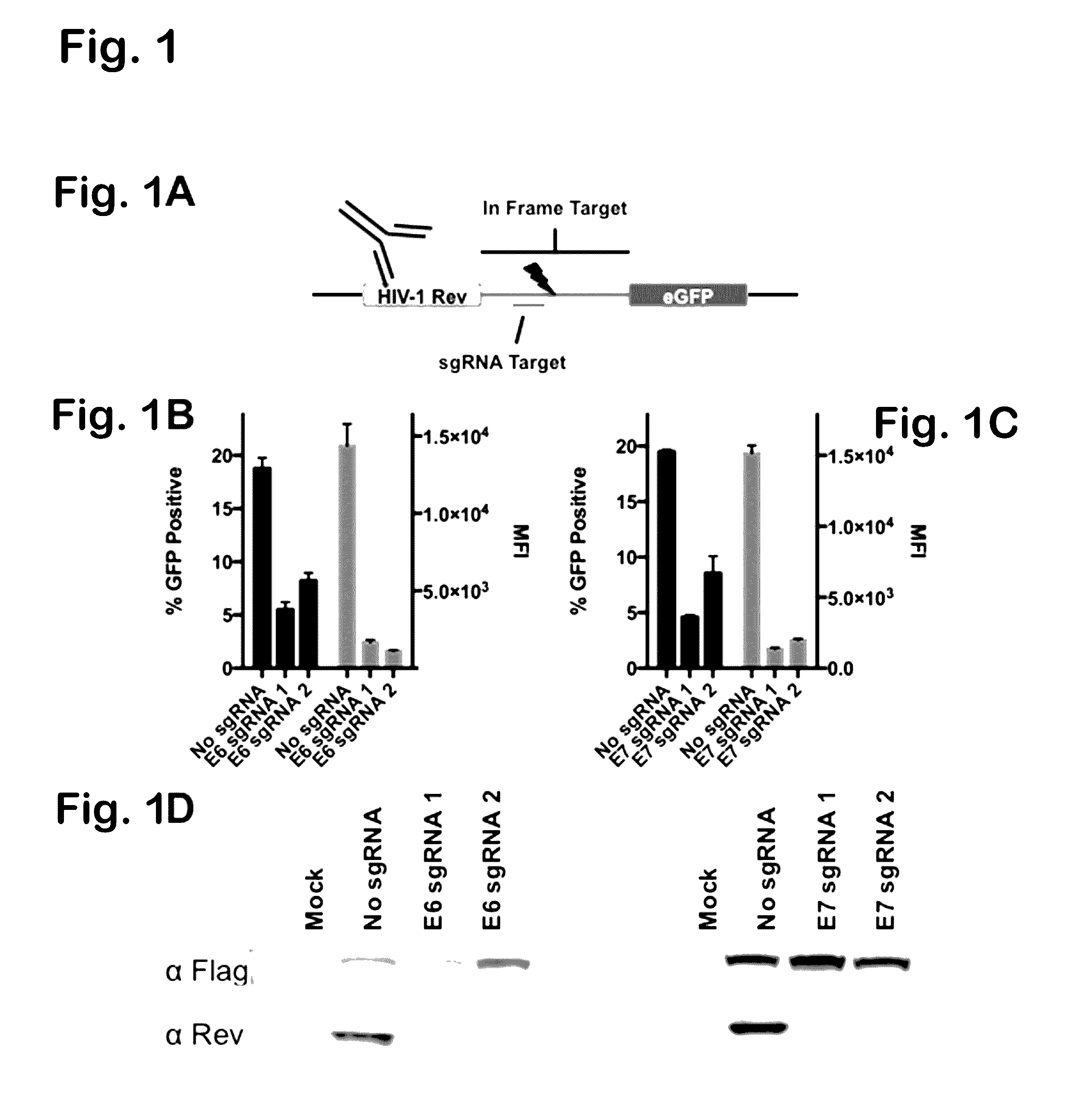
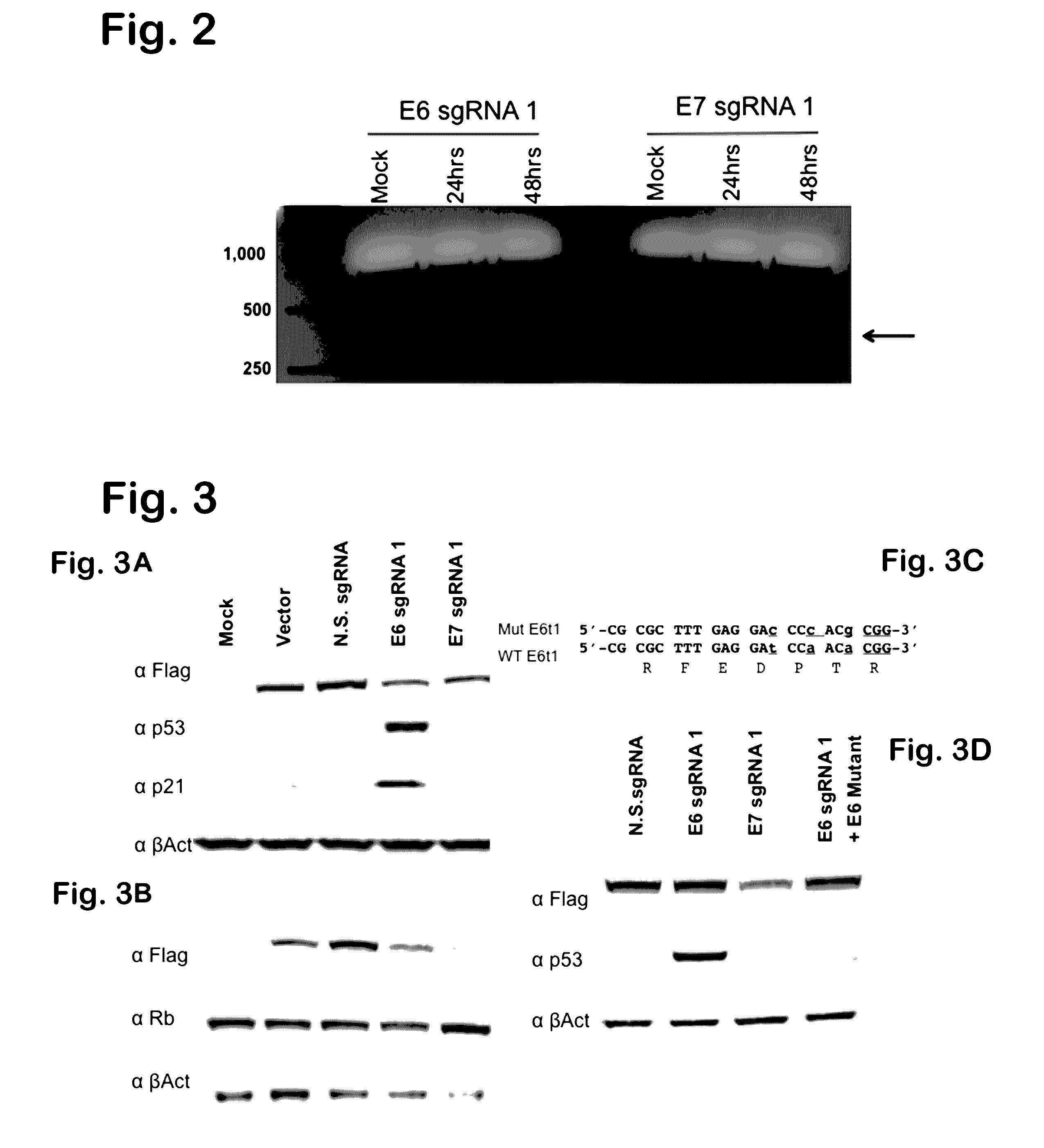
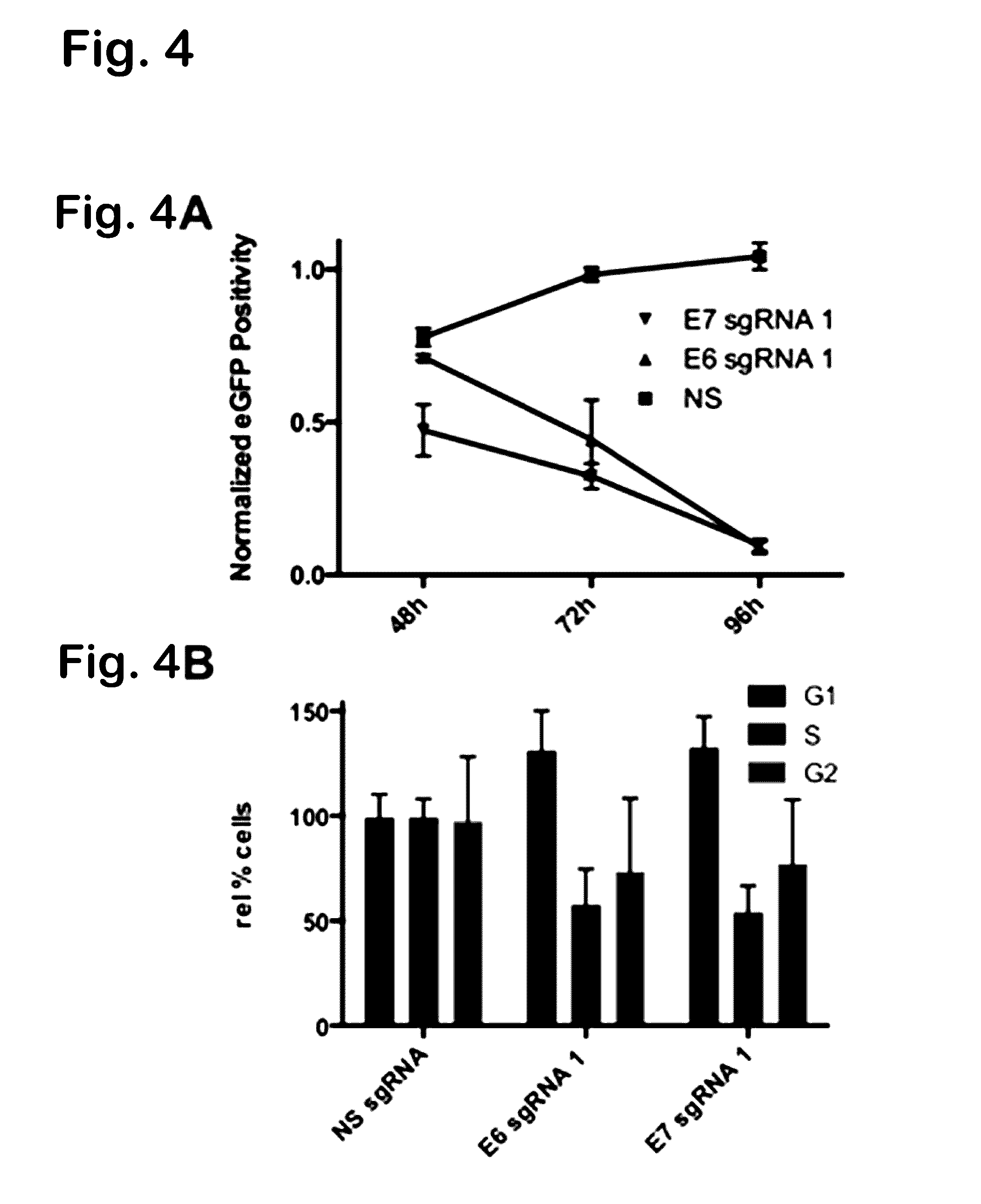
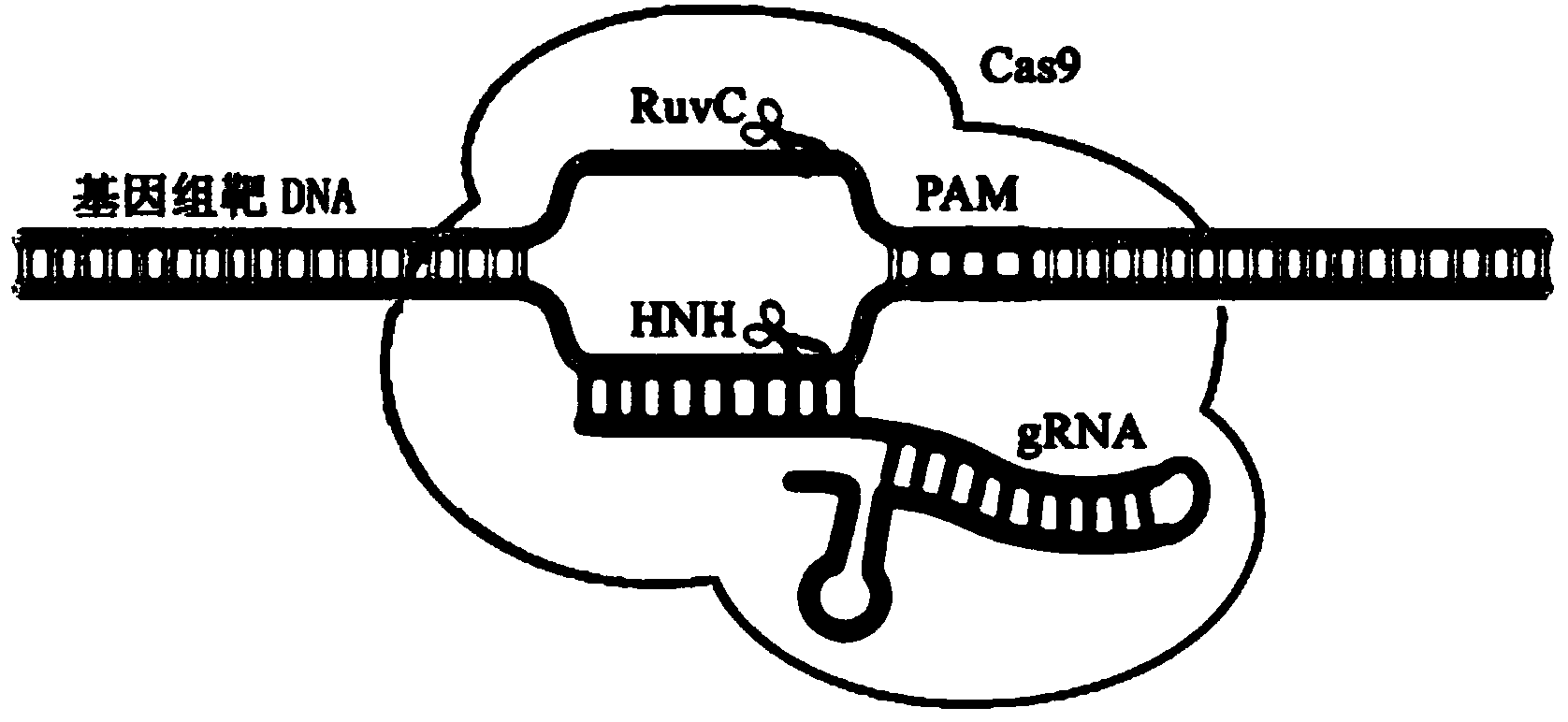
Provided herein are recombinant constructs, vectors and expression cassettes including a first promoter which is suitably a tRNA promoter operably connected to a first polynucleotide encoding a first single guide RNA and a second promoter operably connected to a second polynucleotide encoding a Cas9 polypeptide. The first single guide RNA includes a first portion complementary to a strand of a target sequence of a DNA virus and a second portion capable of interacting with the Cas9 polypeptide. Also provided are codon optimized Staphylococcus aureus derived Cas9 polynucleotides and polypeptides with nuclear localization signals and optionally an epitope tag. Also provided are constructs for production of sgRNAs including a tRNA. Methods of inhibiting viral replication, inhibiting expression of a target sequence from a virus or treating a viral infection or viral induced cancer using the compositions are also provided.
Owner:EMORY UNIVERSITY +2
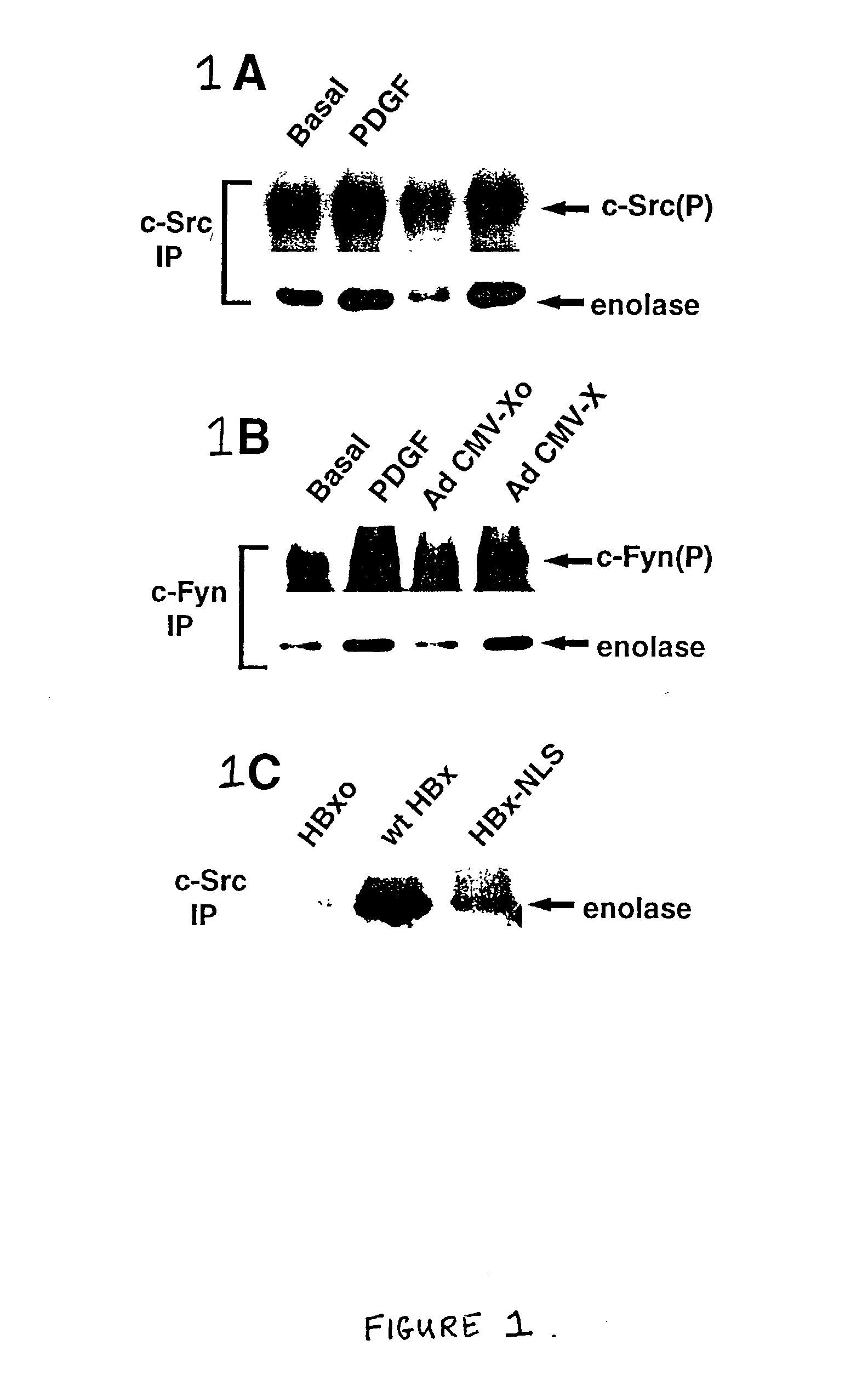
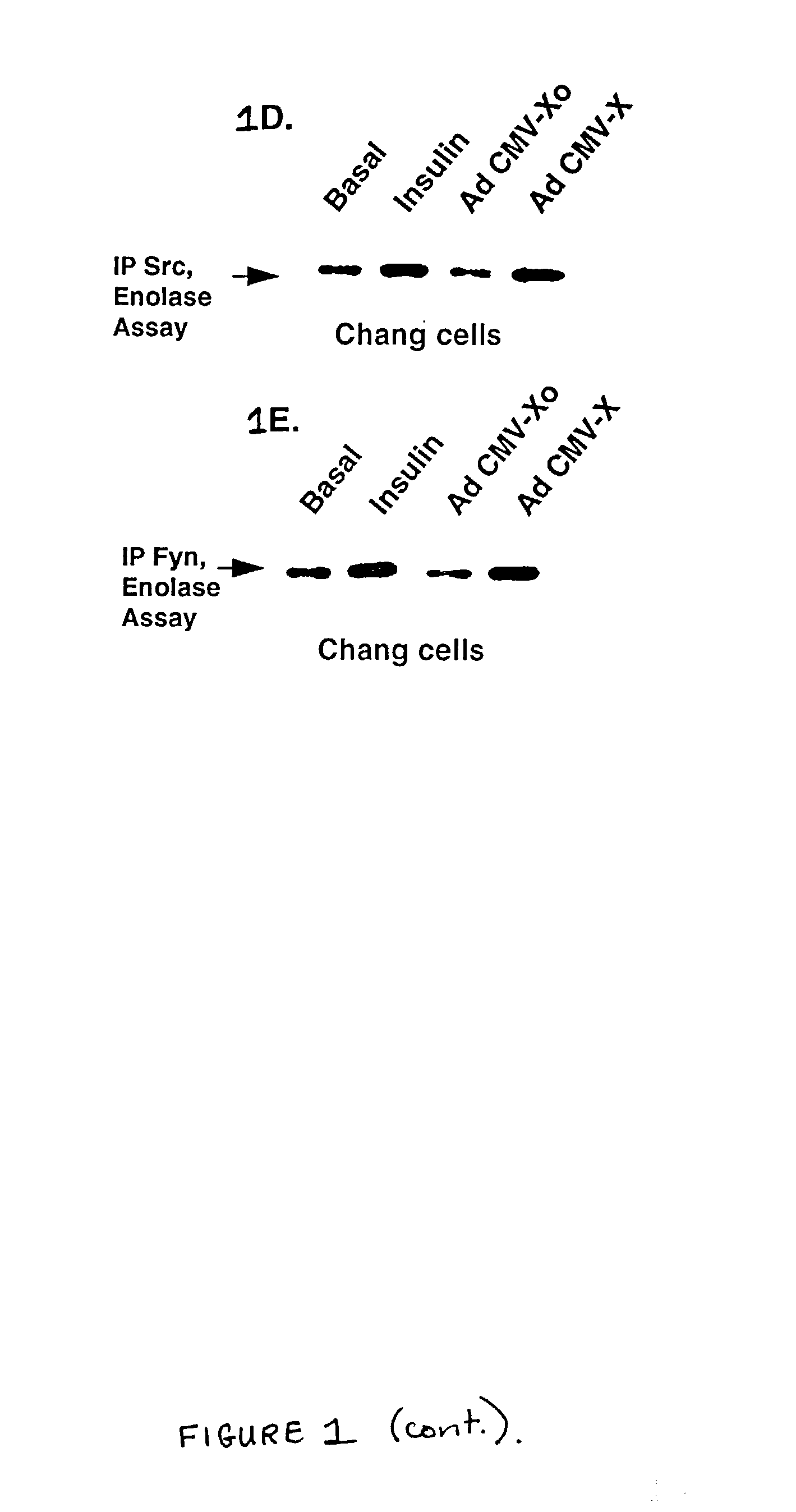
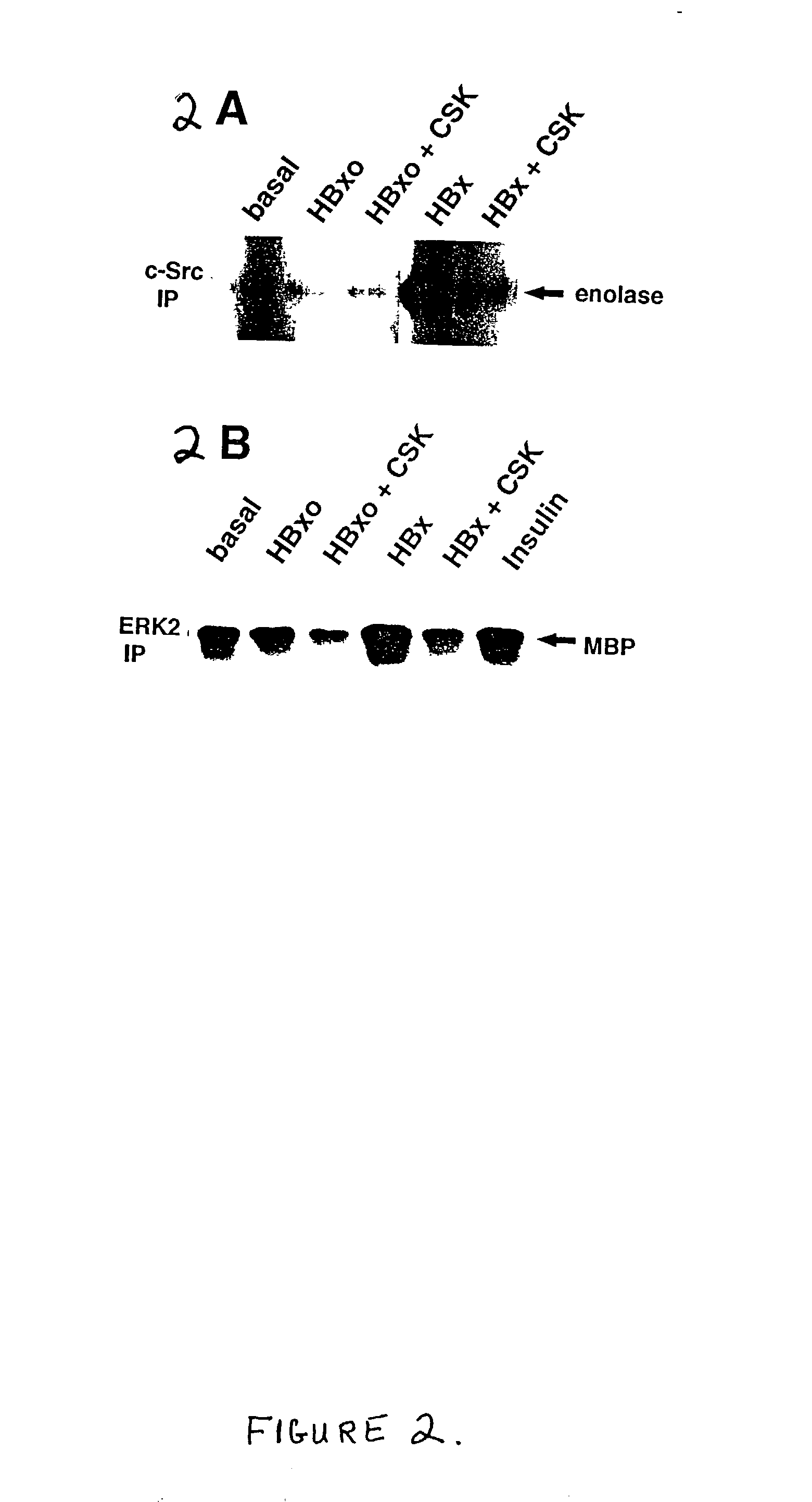
Inhibition of the Src kinase family pathway as a method of treating HBV infection and hepatocellular carcinoma
InactiveUS20030032596A1Highly specific and efficacious methodImprove efficacyBiocidePeptide/protein ingredientsDiseaseHepatocellular carcinoma
Owner:NEW YORK UNIV MEDICAL CENT
Method for co-extracting DNA/RNA (Deoxyribonucleic Acid/Ribonucleic Acid) virus nucleic acid
InactiveCN105925568AImprove throughputHigh-throughput extractionMicrobiological testing/measurementDNA preparationMagnetic beadBiology
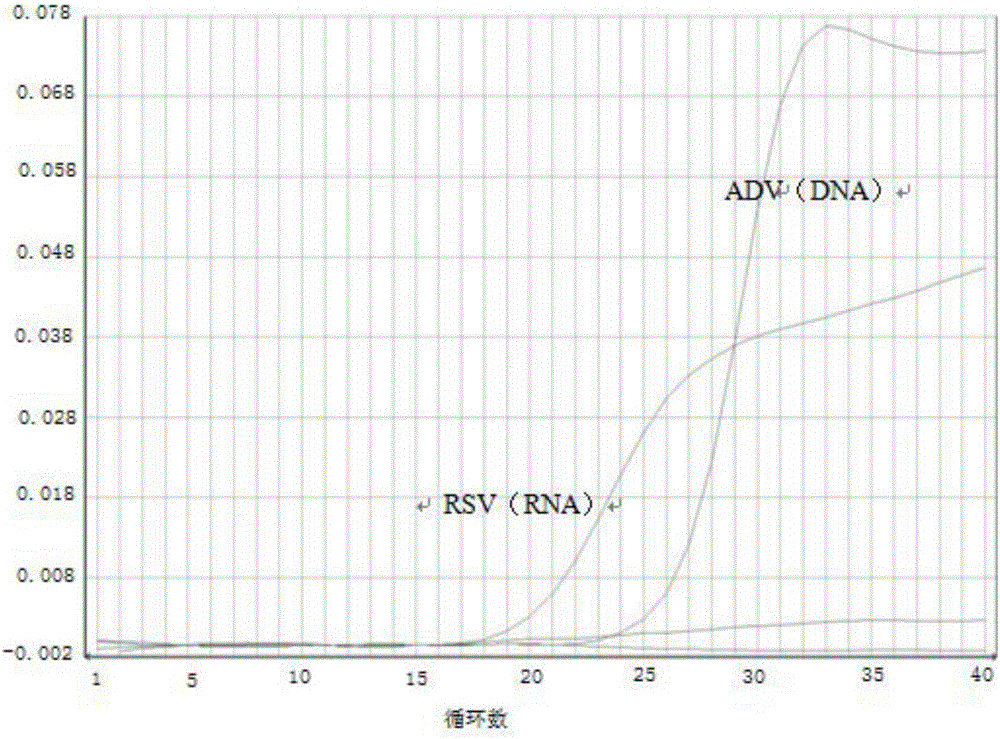
The invention provides a method for co-extracting DNA / RNA (Deoxyribonucleic Acid / Ribonucleic Acid) virus nucleic acid. The method comprises the following steps: cracking a sample, which is diluted with saline, with a guanidine isothiocyanate cracking solution containing an RNA precipitating aid agent; releasing and dissociating DNA or RNA of different nucleic acid viruses in the sample into a cracking solution; adding magnetic beads to form a magnetic bead-nucleic acid compound; washing to obtain a nucleic acid eluting solution. The novel method for co-extruding the DNA and the RNA from a respiratory tract sample is established based on the nano magnetic beads; in a whole process, toxic reagents including phenol, chloroform, beta-mercaptoethanol and the like are not used, and time and labor are saved. The method is used for commonly extracting the DNA and the RNA from the respiratory tract sample and is also applicable to nucleic acid co-extraction of other samples with different types, such as urine and blood; impurities including protein and the like are efficiently removed and the degradation of the nucleic acid is reduced; the aim of detecting DNA viruses and RNA viruses in the same tube at the same time is realized. The method is simple, convenient, rapid and safe, and is particularly suitable for fluorescent quantitative PCR (Polymerase Chain Reaction) detection and the like.
Owner:HANGZHOU FIRST PEOPLES HOSPITAL
Treatment of neoplasms with viruses
The subject invention relates to viruses that are able to replicate and thereby kill neoplastic cells with a deficiency in the IFN-mediated antiviral response, and their use in treating neoplastic disease including cancer and large tumors. RNA and DNA viruses are useful in this regard. The invention also relates to methods for the selection, design, purification and use of such viruses for cancer therapy.
Owner:WELLSTAT BIOLOGICS CORP
Method of simultaneously extracting animal DNA (Deoxyribonucleic Acid) virus and RNA (Ribonucleic Acid) virus nucleic acid in blood serum and double swabs
The invention discloses a method of simultaneously extracting animal DNA (Deoxyribonucleic Acid) virus and RNA (Ribonucleic Acid) virus nucleic acid in blood serum and double swabs. The method comprises the following steps of: carrying out lysis on a to-be-extracted substance by using guanidinium isothiocyanate lysate; adsorbing RNA by a silica gel membrane; removing impure protein by washing liquor I; removing impurities by washing liquor II; carrying out DEPC (Diethylpyrocarbonate) water-washing to remove nucleic acid, wherein the guanidinium isothiocyanate lysate comprises 3M-7M guanidinium isothiocyanate, 0.6%-1.0% TriTon-100, 30mM-50mM Tris-Cl, 5mM-15mM DTT (DL-Dithiothreitol), 60 mu g / mL-90 mu g / mL protease K, 10mM-30mM EDTA (Ethylene Diamine Tetraacetic Acid), and PH of the guanidinium isothiocyanate lysate is 4.3-4.6; the washing liquor I comprises 5M-6M guanidine hydrochloride, 53%-59% absolute ethyl alcohol, 70-90 mu g / mL protease K, and PH of the washing liquor I is 6.4-6.6; the washing liquor II comprises 70%-80% alcohol. The method disclosed by the invention has the advantages of being simple in extracting process, short in period, low in cost, and capable of simultaneously extracting RNA virus and DNA virus nucleic acid in an animal blood serum sample and a double-swab sample.
Owner:安徽华卫集团禽业有限公司
Infection and treatment of neoplasms with vesicular stomatitis virus
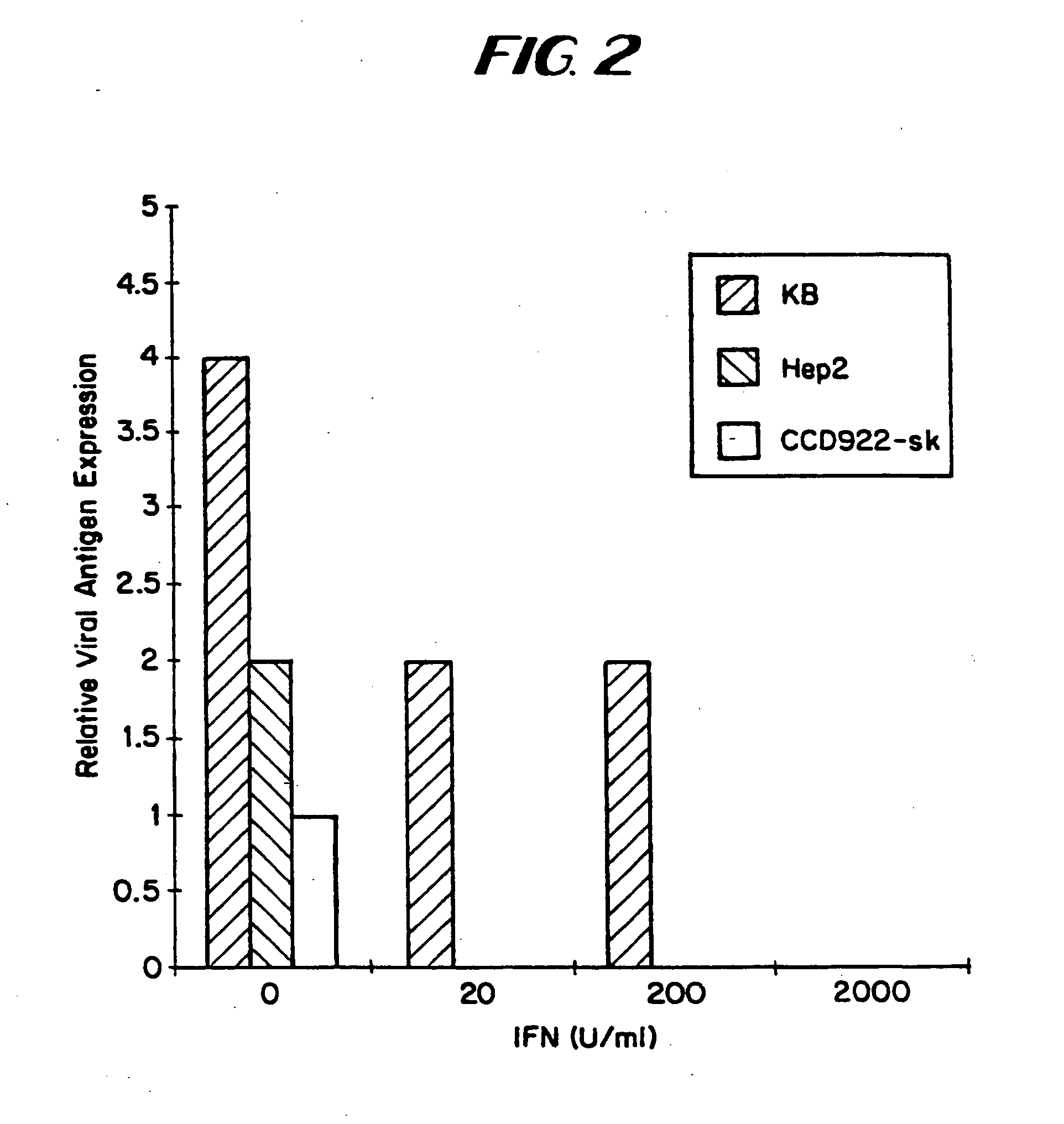
InactiveUS8043612B2Convenient treatmentIncreased interferon sensitivityOrganic active ingredientsVirusesDiseaseDna viral
Owner:WELLSTAT BIOLOGICS CORP
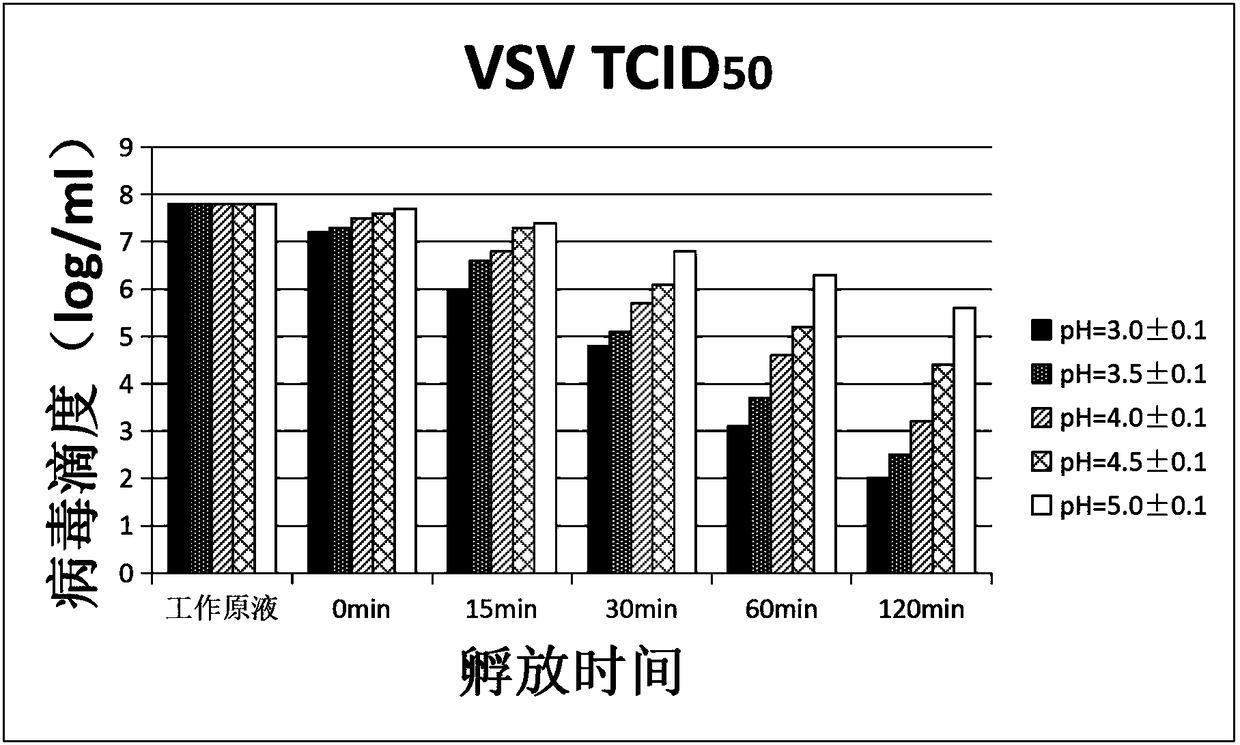
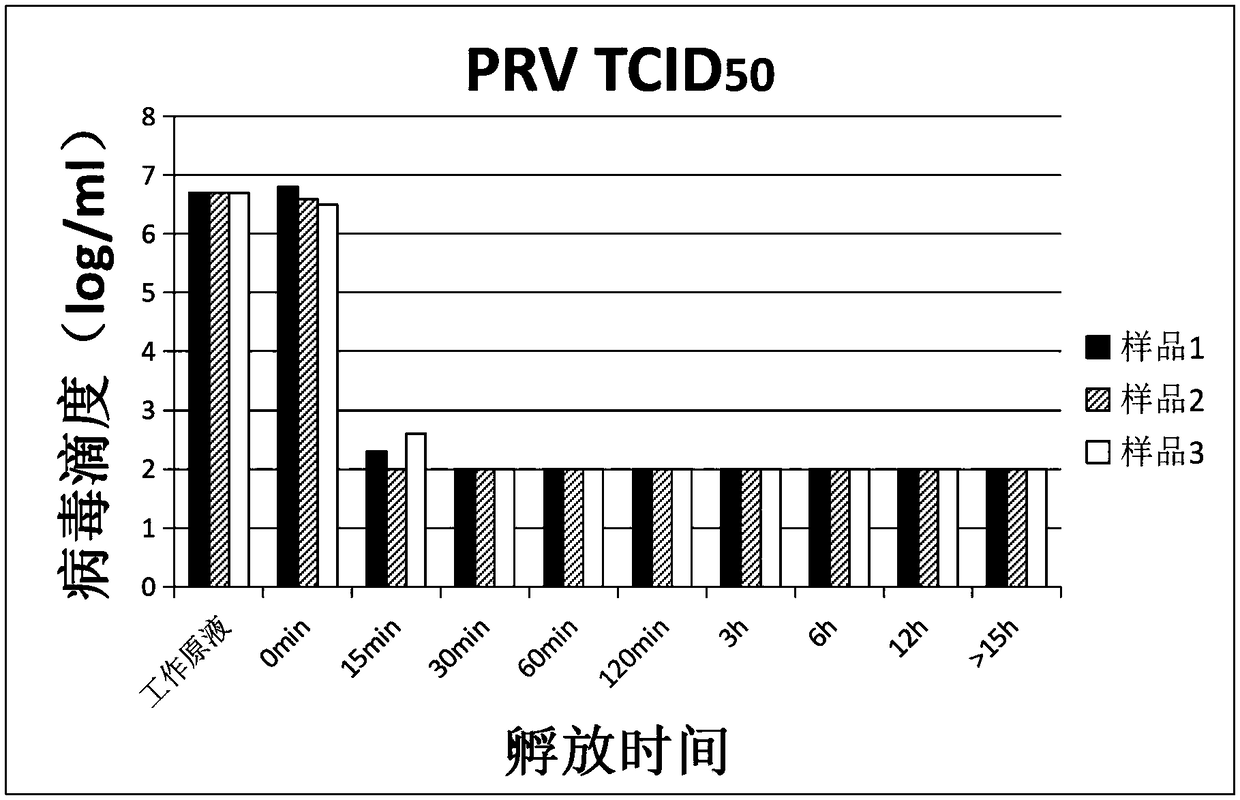
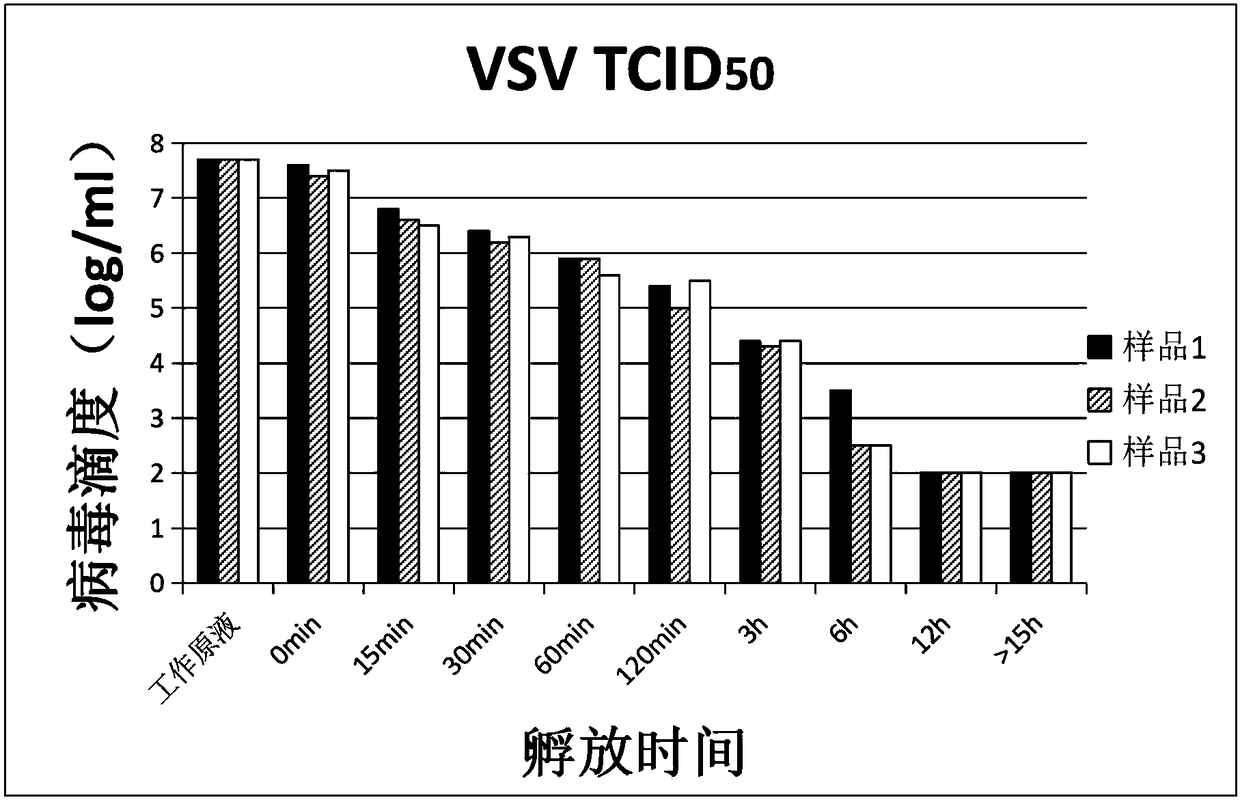
Verification method of low pH incubation virus inactivation
InactiveCN108342368AImprove scalabilitySimple purification processSsRNA viruses negative-senseMicrobiological testing/measurementVirus inactivationValidation methods
The invention relates to a verification method of low pH incubation virus inactivation. The method comprises the steps of selecting indicator viruses and corresponding host cells; amplifying and purifying the indicator viruses; titering virus working primary liquid and samples; optimizing the pH value of virus inactivation verification experiments; estimating the low pH incubation virus inactivation effect. By means of the method, the full process of genetically engineered drug production technology virus inactivation can be simulated in a laboratory, and the amplifying and purifying technology of the indicator viruses is optimized, so that the titer of the working primary liquid of the indicator viruses is increased, the pH value is unified to 7.0, the quality is stable, and subsequent operation is facilitated; meanwhile, the specific technological details like the determination of the optimal incubation pH value and the optimization of the pH adjusting mode of low pH incubation are optimized, and the efficiency and the effectiveness of the inactivation of common DNA viruses, RNA viruses, enveloped viruses and non-enveloped viruses are improved.
Owner:CANVEST WUHAN BIOTECH
Azole nucleosides and use as inhibitors of RNA and DNA viral polymerases
InactiveUS20100129317A1Inhibition is effectivePrevent slippingBiocideSugar derivativesCrimean Congo hemorrhagic fever virusPolymerase L
Azole nucleosides represented by the formulae (I) and (II); wherein A=C or N B═C or N X═H; C1-C6 alkyl, cycloalkyl, alkenyl, cycloalkenyl, alkynyl, aryl, heterocyclo, halogen such as F, Cl, Br and I; OH, NH2, NH—(C1-C6 alkyl, cycloalkyl, aryl or heterocyclo); Z═H; C1-C6 alkyl, cycloalkyl, alkynyl, aryl, heterocyclo, halogen such as F, Cl, Br, I; OH NH2, NH—(C1-C6 alkyl, cycloalkyl, aryl or heterocyclo; E=(CH2)HONHR; n is an interger from 0-6 and more typically 0-3; R1= aryl or heterocyclo; each of W, Y, R is individually selected from the group consisting of H; C1-C6 alkyl, cycloalkyl, alkenyl, cycloalkenyl, alkynyl, aryl, heterocyclo, halogen such as F, Cl, Br, and I; O, OH, Oalkyl, Oaryl, NH2, NH(C1-C6 alkyl, cycloalkyl, aryl or heterocyclo); provided that at least one of W, Y, and R is other than H and wherein both W and Y together can be ═O; and each D individually is OH, Oalkyl, Oaryl, FL and H; pharmaceutically acceptable salts thereof, prodrugs thereof and mixtures thereof are provided. Compounds of this disclosure are useful as inhibitors of viral RNA and DNA polymerases such as, but not limited to, Influenza, hantaan Virus, Crimean Congo hemorrhagic fever virus, hepatitis B, hepatitis C, Polio, Coxsackie A and B, Rhino, Echo, orthopoxvirus (small pox), HIV, Ebola, and West Nile virus polymerases; and especially orthopoxvirus, HIV, and hepatitis B.
Owner:SOUTHERN RES INST & IP +1
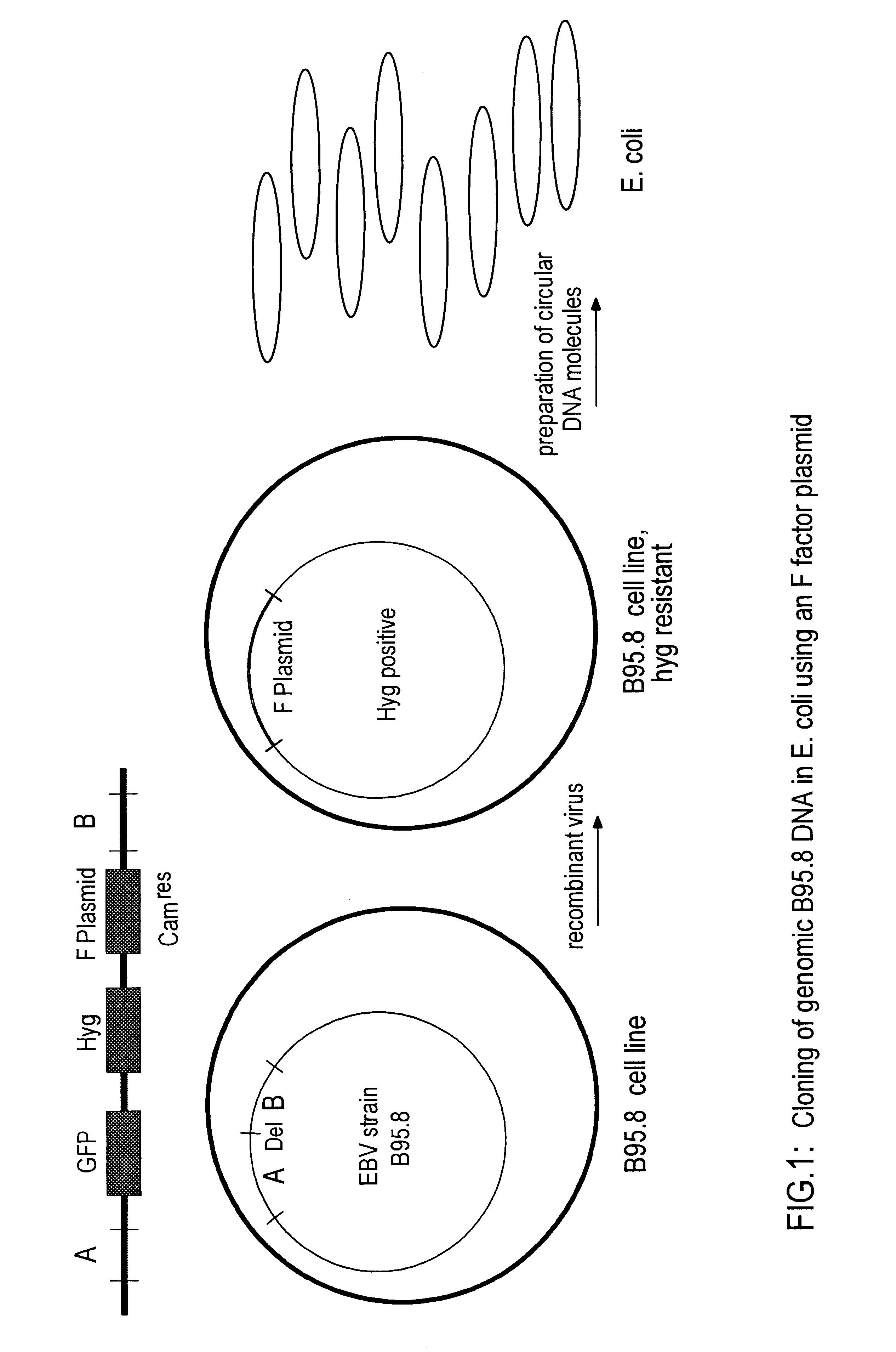
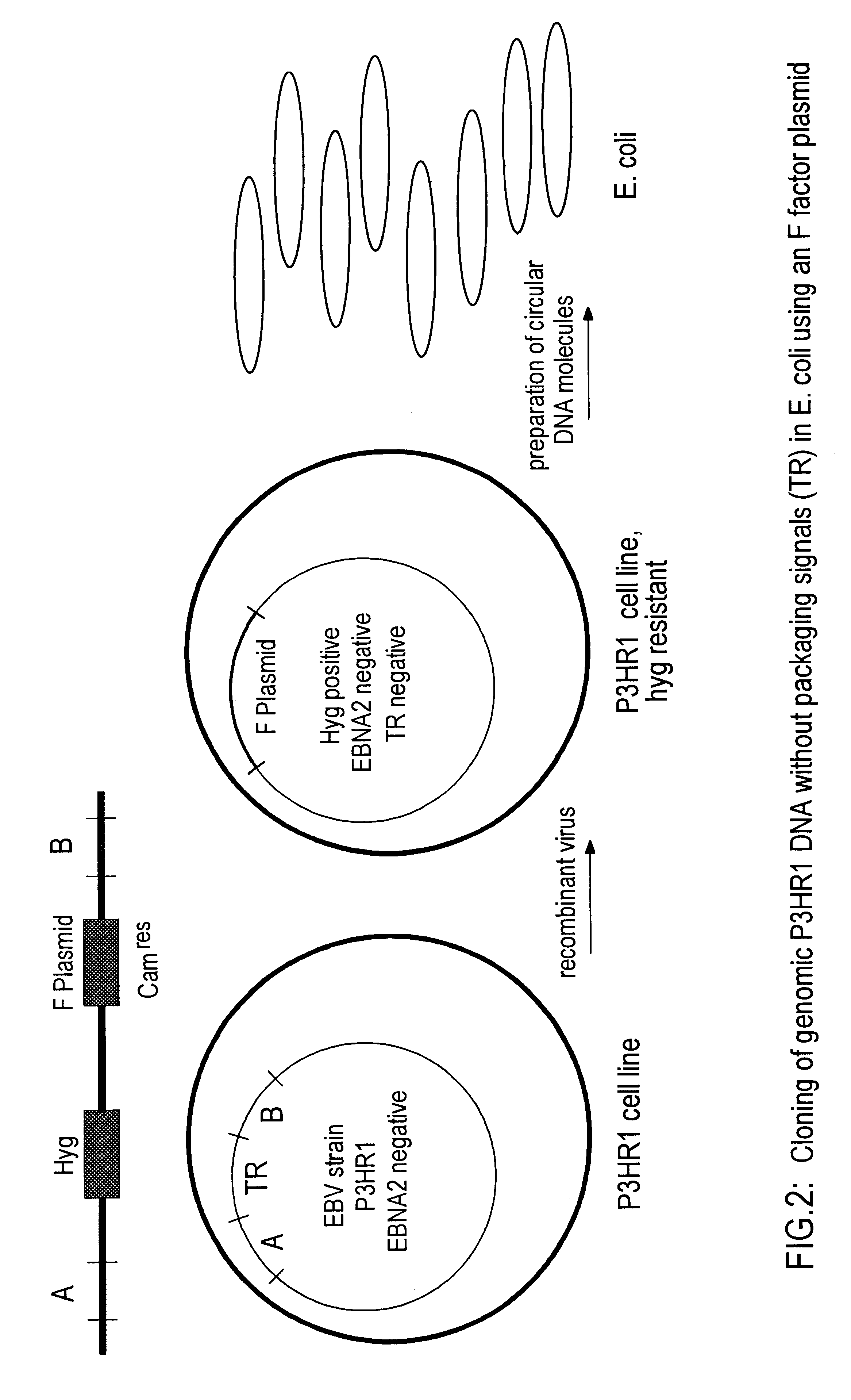
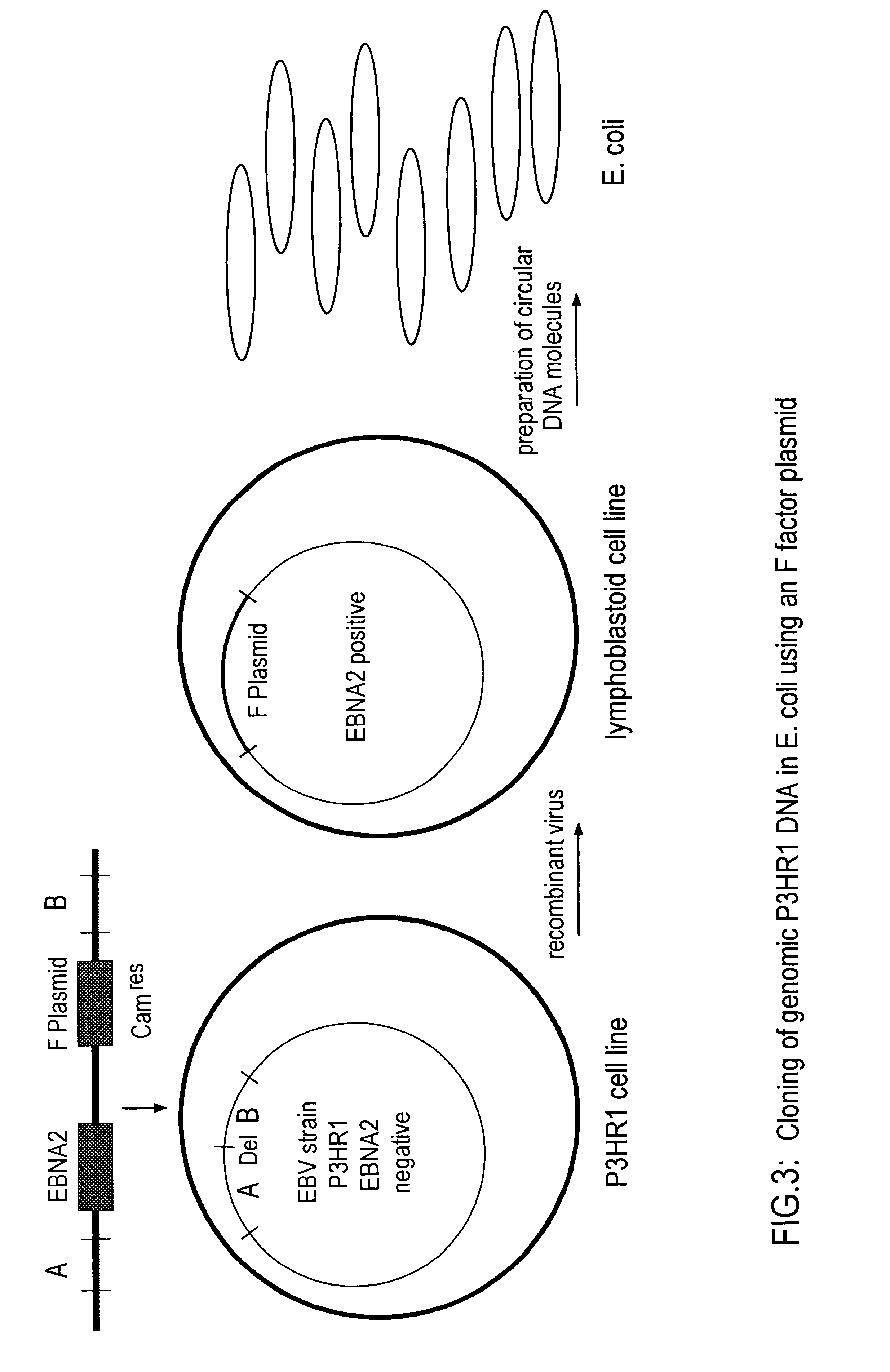
DNA virus vectors and methods for their preparation
InactiveUS6291246B1Improve efficiencyDifficult to controlBacteriaGenetic material ingredientsDna viralVirology
The invention relates to a method for the preparation of DNA virus vectors capable of replication in eukaryotic as well as in prokaryotic cells as well as to DNA virus vectors prepared by this method. Preferably, the method is used to prepare Epstein-Barr virus vectors.
Owner:GSF FORSCHUNGSZENT FUR UMWELT & GESUNDHEIT
Dual SYBR Green I real-time fluorescence PCR detection primer and method for porcine pseudorabies virus and porcine circovirus type 2
InactiveCN102071256AEasy to identifyEasy diagnosisMicrobiological testing/measurementMicroorganism based processesFluorescencePorcine circovirus
The invention discloses a dual SYBR Green I real-time fluorescence polymerase chain reaction (PCR) detection primer and a dual SYBR Green I real-time fluorescence PCR detection method for porcine pseudorabies virus and porcine circovirus type 2. The primer is designed and synthesized, and the dual SYBR Green I real-time fluorescence PCR detection method for the porcine pseudorabies virus and the porcine circovirus type 2 by using the primer comprises the following steps of: extracting deoxyribose nucleic acid (DNA) of a sample; and detecting the sample by using an SYBR Green I real-time fluorescence PCR reaction system and an SYBR Green I real-time fluorescence PCR amplification program. By the primer and the method, two viruses, namely the porcine pseudorabies virus and the porcine circovirus type 2 can be detected simultaneously, and porcine parvovirus, classical swine fever virus, porcine reproductive and respiratory syndrome virus and swine influenza virus cannot be detected. The primer and the method have the characteristics of higher sensitivity, repeatability and stability, and contribute to the identification and the diagnosis of pregnant sow reproductive disturbance virus disease.
Owner:HENAN AGRICULTURAL UNIVERSITY
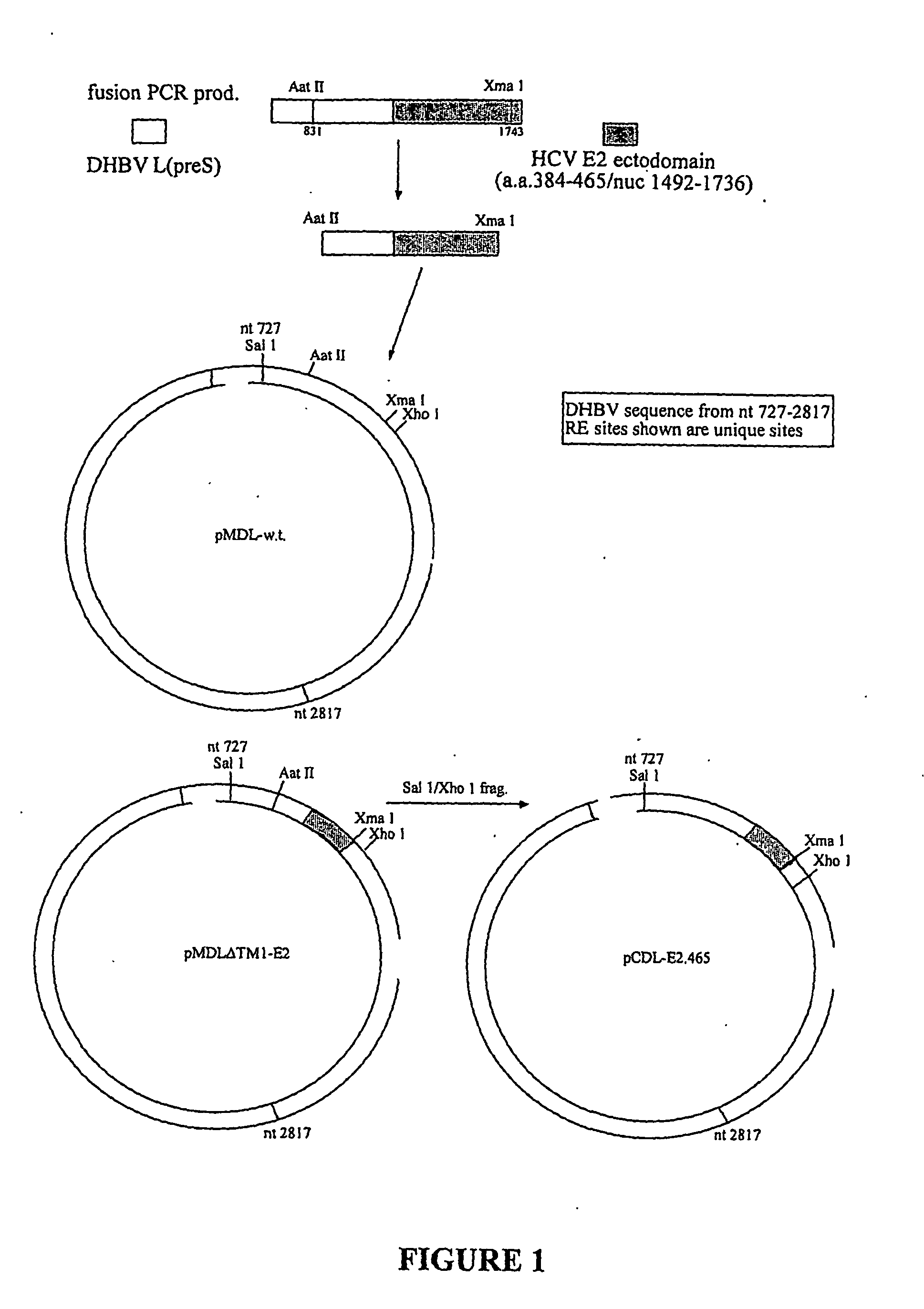
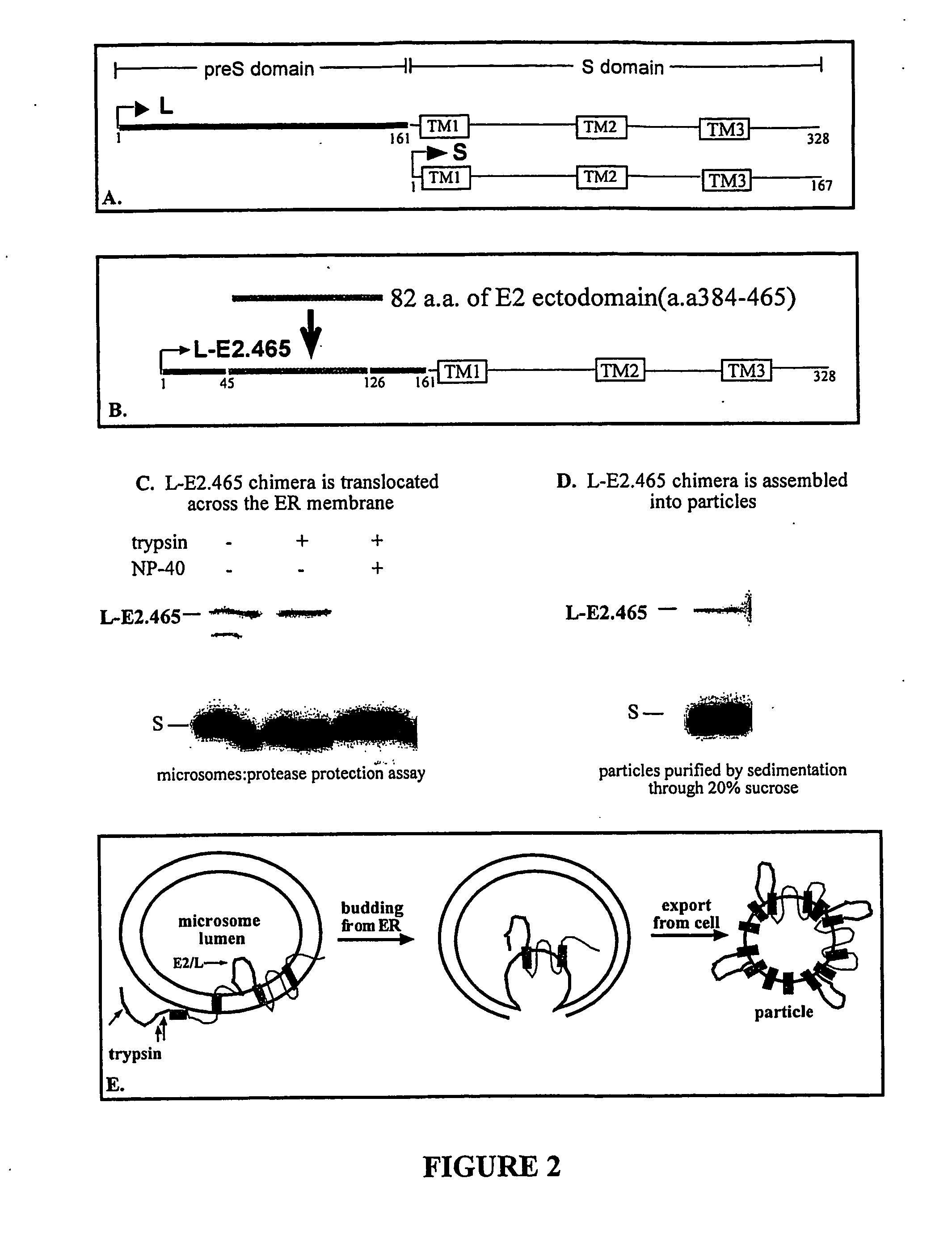
Viral vectors expressing fusion of viral large envelope protein and protein of interest
The present invention provides a virus-like particle (VLP) comprising i) a polypeptide comprising a polypeptide of interest (POI) and at least a particle-associating portion of a large envelope (L) polypeptide of an avian hepadnavirus or a functional derivative or homolog thereof, and ii) a small envelope (S) polypeptide of an avian hepadnavirus or a functional derivative or homolog thereof. By introducing one or more POIs into the L polypeptide, the POI is translocated along with L into a particle structure made up primarily of S polypeptide. The present invention furthermore provides methods for producing a recombinant virus-like particle.
Owner:HEPGENICS
Treatment of neoplasms with viruses
InactiveUS20090081161A1Convenient treatmentIncreased interferon sensitivityOrganic active ingredientsVirusesDiseaseAnti viral response
The subject invention relates to viruses that are able to replicate and thereby kill neoplastic cells with a deficiency in the IFN-mediated antiviral response, and their use in treating neoplastic disease including cancer and large tumors. RNA and DNA viruses are useful in this regard. The invention also relates to methods for the selection, design, purification and use of such viruses for cancer therapy.
Owner:WELLSTAT BIOLOGICS CORP
Methods of producing a library and methods of selecting polynucleotides of interest
InactiveUS20060160129A1Improve efficiencyVirusesMicrobiological testing/measurementNucleotideDna viral
Owner:UNIVERSITY OF ROCHESTER
Novel plant DNA virus carrier and its use
InactiveCN1425772AEasy accessClear functionMicrobiological testing/measurementGenetic engineeringViral FunctionForeign protein
The plant DNA virus carrier includes the infectious carrier of Chinese tomato yellowing curly leaf virus DNA-A and DNA-beta, derived plant gene silencing carrier and virus expressing carrier. The present invention also provides one new kind of satellite DNA molecule constructing plant DNA virus carrier and it has at least 72% homology with any one sequence of SEQ ID No.1-12. The present invention also provides the use and method of utilizing plant DNA virus carrier in virus functional genome research, plant functional genome research and foreign protein expression. The present invention provides one important platorm for researching virus genomee structure, function and expression and regulation mechanism and the interaction between virus and plant; provides one excellent tool for researching plant functional gene; and establishing one technological platform for using plant as bioreactor expressing foreign protein.
Owner:ZHEJIANG UNIV
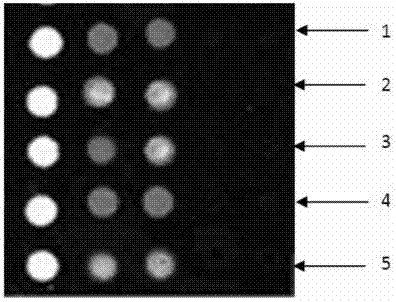
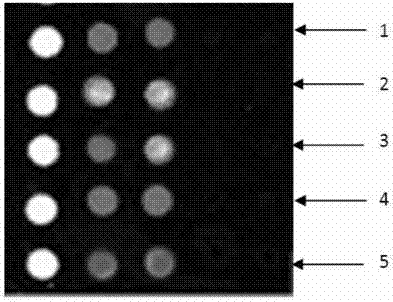
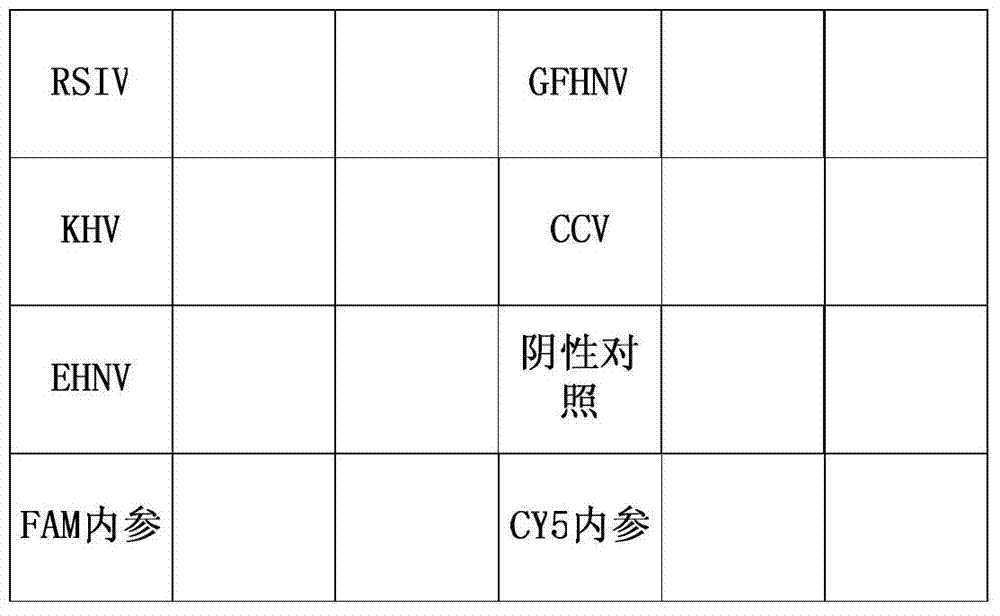
Gene chip kit for aquatic animal DNA (deoxyribonucleic acid) virus detection, and preparation method and application thereof
ActiveCN103757133ALow detection costExpand the scope of detectionMicrobiological testing/measurementNucleotideAquatic animal
The invention relates to a gene chip for detecting aquatic animal DNA (deoxyribonucleic acid) viruses, which comprises a solid-phase carrier and an aquatic animal DNA virus molecule beacon probe fixed to the solid-phase carrier, wherein the molecule beacon probe comprises nucleotide sequences disclosed as SEQ ID NO.3-7, the 5' end of the nucleotide sequences disclosed as SEQ ID NO.3-7 is connected with FAM fluorescein, and the 3' end is connected with a Dabcyl quenching group. The invention also relates to a kit for detecting aquatic animal DNA viruses, and a preparation method and application thereof. The gene chip and kit for detecting aquatic animal DNA viruses can implement one-step quick detection on all infected aquatic animal DNA viruses, have higher sensitivity, and reduce the detection cost for individual samples.
Owner:SHENZHEN AUDAQUE DATA TECH
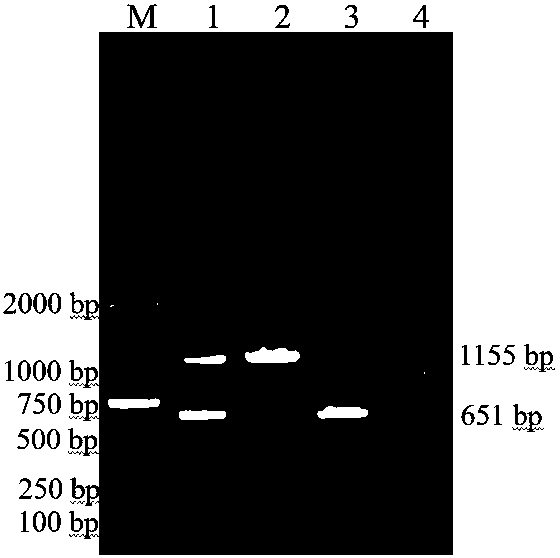
Duplex PCR (polymerase chain reaction) detection primer and kit for quickly distinguishing porcine circoviruses type 2 and type 3
InactiveCN108531656AEasy to operateQuick checkMicrobiological testing/measurementMicroorganism based processesDuplex pcrSterile water
The invention belongs to the technical field of animal virology and molecular biology and discloses a duplex PCR (polymerase chain reaction) detection primer and kit for quickly distinguishing porcinecircoviruses type 2 and type 3. The kit comprises primer sequences shown as SEQ ID1-4, 2*F8 Fastlong PCR MasterMix, a cDNA template and sterile water. The duplex PCR detection kit for quickly distinguishing the porcine circoviruses type 2 and type 3 has advantages that by duplex PCR amplification, two types of DNA viruses similar in lesion can be detected in one time, the total detection time iscontrolled to be about two hours, a detection method is easy in operation, convenient and quick, and quick detection can be realized.
Owner:GUANGXI VETERINARY RES INST
Site-specific modification and screening method for specific DNA (deoxyribonucleic acid) viral genome
ActiveCN103757053AAchieve expression efficiencyEasy to filterViruses/bacteriophagesVector-based foreign material introductionRandom mutationHomologous sequence
The invention provides a site-specific modification and screening method for a specific DNA (deoxyribonucleic acid) viral genome. The method comprises the following steps:1, establishing site-specific cleavage single-strand and duplex nuclease systems and a homologous sequence; 2, introducing cytocidal infection DNA viruses of the site-specific cleavage single-strand nuclease system and the homologous sequence, and collecting P1 progeny viruses after cells are pathologically changed; 3, introducing cytocidal infection P1 progeny viruses of the site-specific cleavage duplex nuclease system, collecting P2 progeny viruses after the cells are pathologically changed, and separating and purifying the P2 progeny viruses to obtain target progeny viruses, wherein the step 2 is a viral genome homologous recombination step, and the viral genome homologous recombination efficiency is improved by about hundreds of thousands of times under the condition of not introducing random mutation; the step 3 is a specific amplification step, and specific mutation viruses in the progeny viruses are further increased about 10 times. According to the method, mutation types can be effectively controlled, and recombinant viruses with specific mutation, deficiency or intercalation can be rapidly and conveniently screened.
Owner:INST OF MEDICAL BIOLOGY CHINESE ACAD OF MEDICAL SCI
Treatment of neoplasms with RNA viruses
InactiveUS7780962B2Convenient treatmentSsRNA viruses negative-senseBiocideDiseaseAnti viral response
The subject invention relates to viruses that are able to replicate and thereby kill neoplastic cells with a deficiency in the IFN-mediated antiviral response, and their use in treating neoplastic disease including cancer and large tumors. RNA and DNA viruses are useful in this regard. The invention also relates to methods for the selection, design, purification and use of such viruses for cancer therapy.
Owner:WELLSTAT BIOLOGICS CORP
Transcriptome sequencing method for microviridae and geminivirus viral genome splice
PendingCN107475449AAccurate identificationAutomate quicklyMicrobiological testing/measurementMicroorganism based processesDna viralTranscriptome Sequencing
The invention provides a transcriptome sequencing method for microviridae and geminivirus viral genome splice. The method comprises the three steps of performing transcriptome sequencing on an infected DNA virus sample, comparing and splicing viral sequences and filling a blank area of the viral genome sequence. According to the method provided by the invention, the whole genome sequence of the banana bunchy top virus can be acquired and the other microviridae or geminivirus viral genomes can be spliced so as to acquire the whole genome sequence of the virus. The invention firstly reports the method for sequencing the spliced DNA viral genome in high throughput by utilizing transcriptome at home and abroad and fills the blank of the field at home and abroad. The method provided by the invention can be used for identifying and detecting DNA virus.
Owner:INST OF TROPICAL BIOSCI & BIOTECH CHINESE ACADEMY OF TROPICAL AGRI SCI
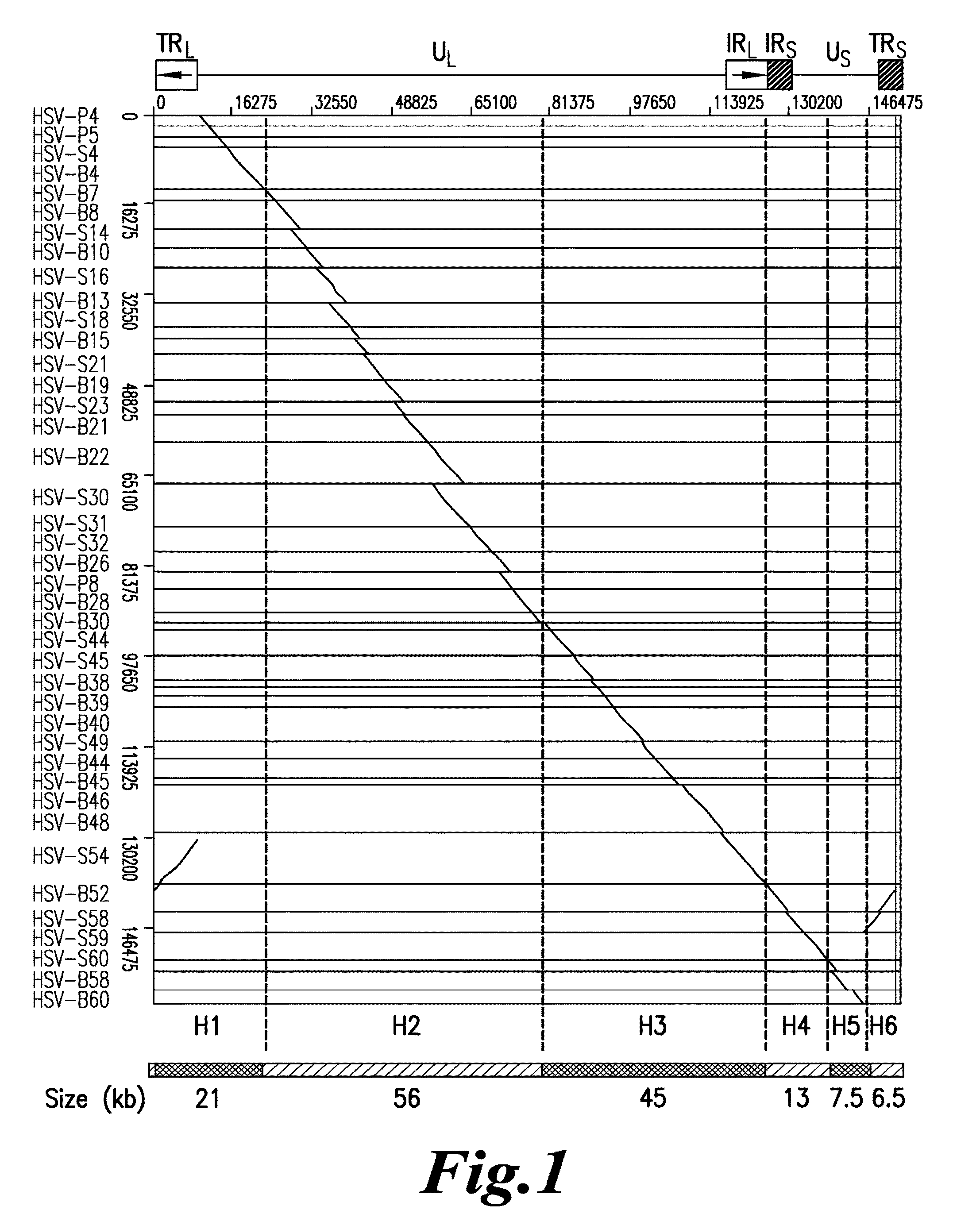
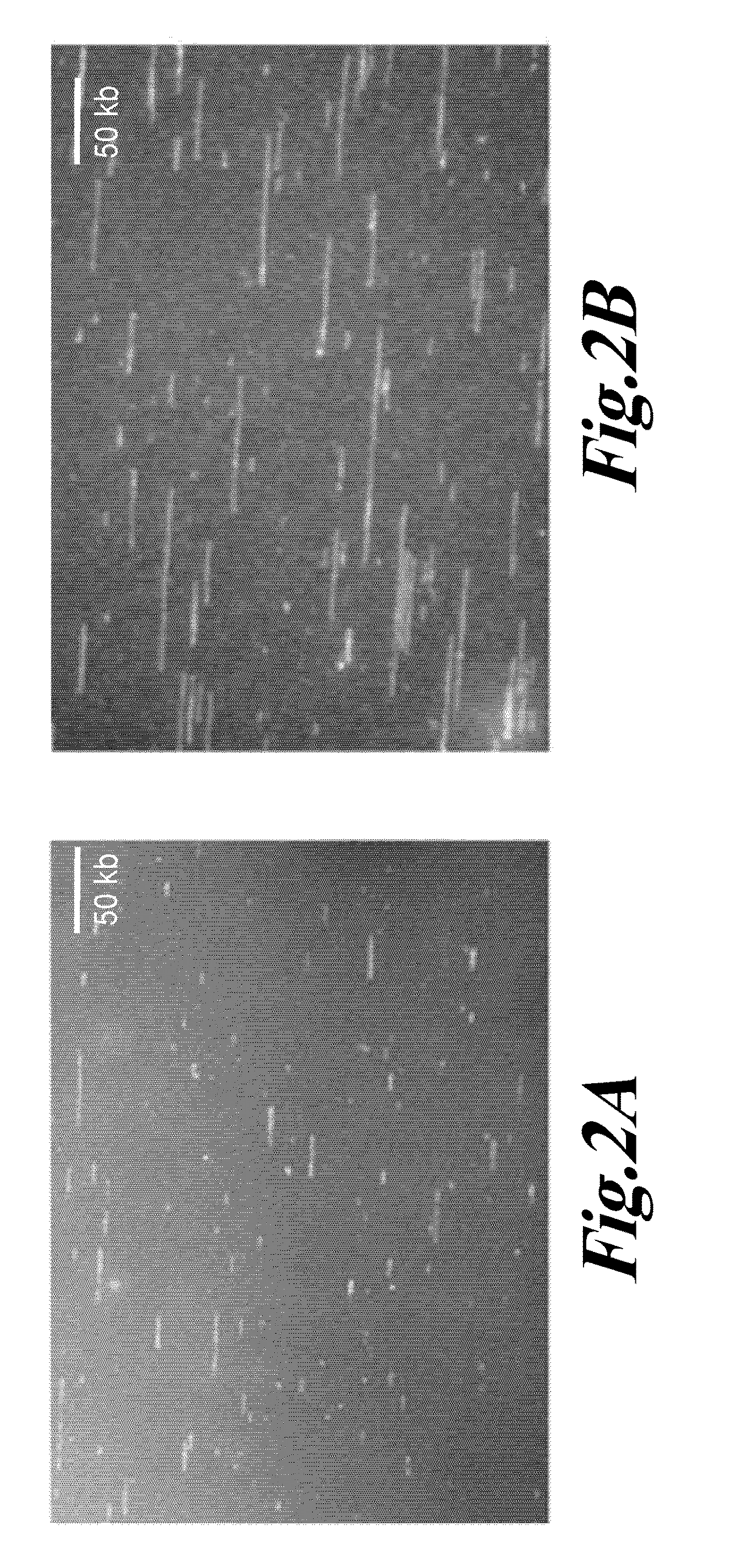
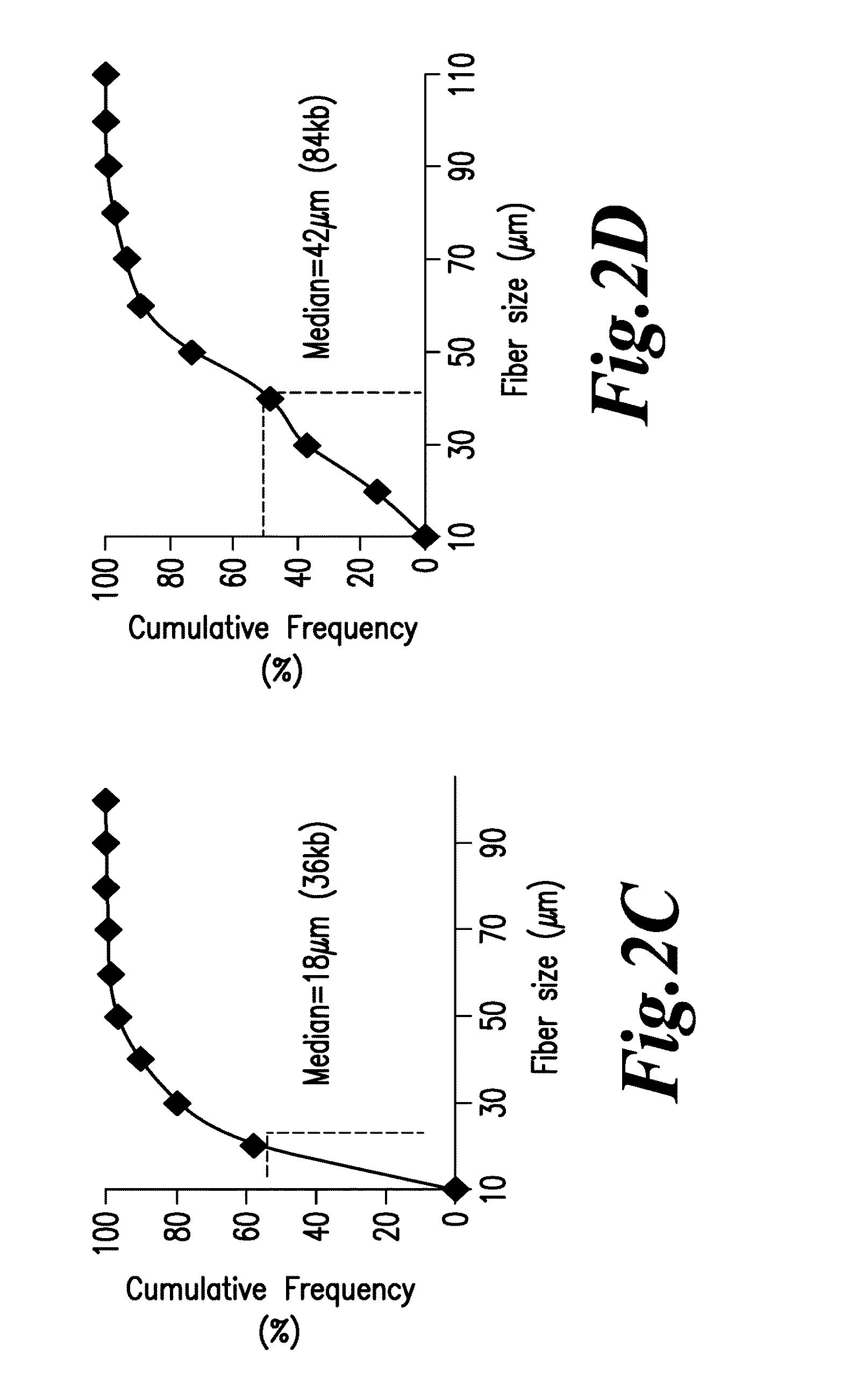
Diagnosis of viral infections by detection of genomic and infectious viral DNA by molecular combing
InactiveUS20160047006A1Reliable and simple and fast and inexpensive to detectNot require multiple time-consume and expensive procedureMicrobiological testing/measurementAntiviralsSpecial designNucleic Acid Probes
A method for detecting in vitro the presence of a genome of a DNA virus or a viral derived DNA in an infected eukaryotic cell, tissue or biological fluid using Molecular Combing or other nucleic acid stretching methods together with probes, especially nucleic acid probes, having a special design. A method for monitoring in vitro the effects of anti-viral treatment by following the presence of genomic viral or viral derived DNA polynucleotides in a virus-infected cell, tissue or biological fluid. Detection of an infectious form of a virus using Molecular Combing and DNA hybridization. A kit comprising probes used to carry out these methods and a composition comprising the probes.
Owner:GENOMIC VISION
Dual SYBR Green I real-time fluorescence PCR detection primer and method for porcine parvovirus and porcine circovirus type 2
InactiveCN102071257AStrong specificityGood repeatabilityMicrobiological testing/measurementMicroorganism based processesAgricultural scienceFluorescence
The invention discloses a dual SYBR Green I real-time fluorescence polymerase chain reaction (PCR) detection primer and a dual SYBR Green I real-time fluorescence PCR detection method for porcine parvovirus and porcine circovirus type 2. The primer is designed and synthesized, and the dual SYBR Green I real-time fluorescence PCR detection method for the porcine parvovirus and the porcine circovirus type 2 by using the primer comprises the following steps of: extracting deoxyribose nucleic acid (DNA) of a sample; and detecting the sample according to an SYBR Green I real-time fluorescence PCR reaction system and an SYBR Green I real-time fluorescence PCR amplification program. By the primer and the method, two viruses, namely the porcine parvovirus and the porcine circovirus type 2 can be detected simultaneously, and porcine pseudorabies virus, classical swine fever virus, porcine reproductive and respiratory syndrome virus and swine influenza virus cannot be detected. The primer and the method have the characteristics of higher specificity, repeatability and sensitivity.
Owner:HENAN AGRICULTURAL UNIVERSITY
Kit for synchronous detection of twenty three respiratory pathogens and detection method thereof
ActiveCN110578017AShort ampliconReduced Chances of ContaminationMicrobiological testing/measurementMicroorganism based processesReverse transcriptaseBiology
The invention discloses a kit for synchronous detection of twenty three respiratory pathogens and a detection method thereof. The kit comprises: a transcription and amplification primer, 2.5 x Reaction MIX, DEPC water, a hot-start tap enzyme and a reverse transcriptase, wherein the transcription and amplification primer includes primer sequences of twenty three respiratory pathogens including 17 RNA viruses, 2 DNA viruses and 4 respiratory tract pathogenic bacteria, and the gene sequences are as shown in SEQ ID NO.1-NO.44. After pathogen samples are extracted, transcription and amplification are completed through single-tube reaction of the kit, capillary electrophoresis detection is performed, and pathogen categories are determined through software analysis of results. The technical scheme has the advantages of strong specificity, high sensitivity, fast detection speed, completion of transcription amplification by a single tube, and simple and convenient operation.
Owner:深圳市百迈生命科学有限公司 +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com