Transcriptome sequencing method for microviridae and geminivirus viral genome splice
A technology of transcriptome sequencing and viral genome, applied in the field of genome sequencing and bioinformatics, can solve the problems of time-consuming and laborious, large homology differences, and inability to obtain the whole genome sequence of new viruses well, reaching the scope of application wide effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Both Dwarfviridae and Geminiviridae DNA viruses produce RNA from the non-coding regions:
[0044] Dwarf Viridae:
[0045] Banana bunchy top virus (BBTV) is the causative agent of banana bunchy top virus (BBTD), and its genome consists of at least six circular ssDNA components ranging in size from 1.0 to 1.1 kb. In 2015, the International Committee on Taxonomy of Viruses assigned it to the genus Babuvirus of the family Nanaviridae and became a representative member of the genus. In this example, taking banana bunchy top virus as an example, it is confirmed that the non-coding regions of Dwarfviridae members can produce RNA, and the steps are as follows:
[0046] (1) Using the extracted total RNA of the B2 sample as a template, after being treated with DNase I, the first-strand cDNA was synthesized using the ReverseTranscriptase M-MLV reverse transcription kit;
[0047] (2) Then design the following primer sequences according to the DNA-R non-coding region (SEQ ID NO.1)...
Embodiment 2
[0059] A method for sequencing virus genomes suitable for Dwarfviridae and Geminiviridae, comprising the following steps (this embodiment takes the banana sample infected by BBTV as an example to illustrate the method):
[0060] (1) Using a commercial TRIzol Reagent kit, extract total RNA from infected BBTV banana leaves (B2);
[0061] (2) Adopt commercialization Ultra TM RNA Library Prep Kit for (NEB, USA) kit: Firstly, mRNA is enriched by magnetic beads with Oligo(dT), and the mRNA is broken into short fragments; then the short fragment mRNA is used as a template to reverse transcribe into the first-strand cDNA; The kit further synthesizes double-stranded cDNA, that is, a cDNA library, which can be used for high-throughput sequencing on a computer (Illumina HiSeqTM 2000 high-throughput sequencing technology platform).
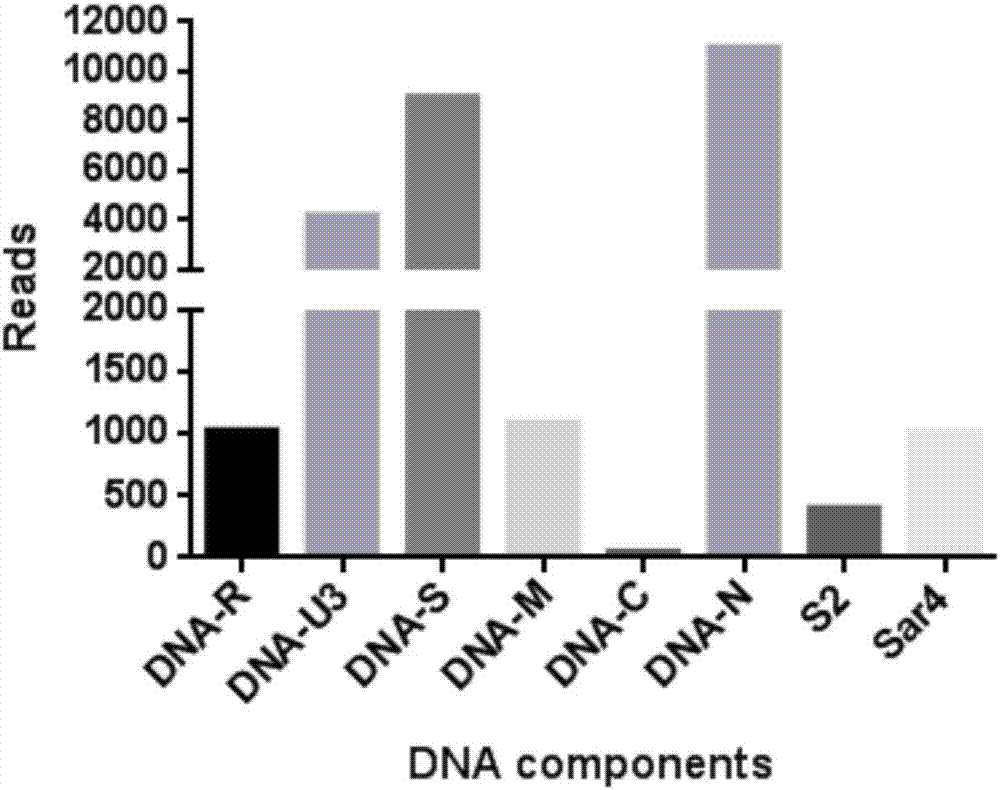
[0062] (3) Run Bowtie software (default parameters), compare the transcriptome sequencing fragments in step (2) with the nucleotide sequences of all kn...
Embodiment 3
[0080] Supplement to the gap area:
[0081] In this embodiment, PCR supplementation is performed on the two blank regions of the DNA-N component, and the steps are as follows:
[0082] (1) Using the extracted total RNA of the B2 sample as a template, use the Reverse Transcriptase M-MLV reverse transcription kit to synthesize the first-strand cDNA, and then design the following primer sequences for PCR amplification:
[0083] Forward primer DNA-N 23F: 5'-CCAGCGCTCGGGACGGG-3';
[0084]Reverse primer DNA-N 878R: 5'-GCCCAATCAACCCCCGATTCTTC-3'.
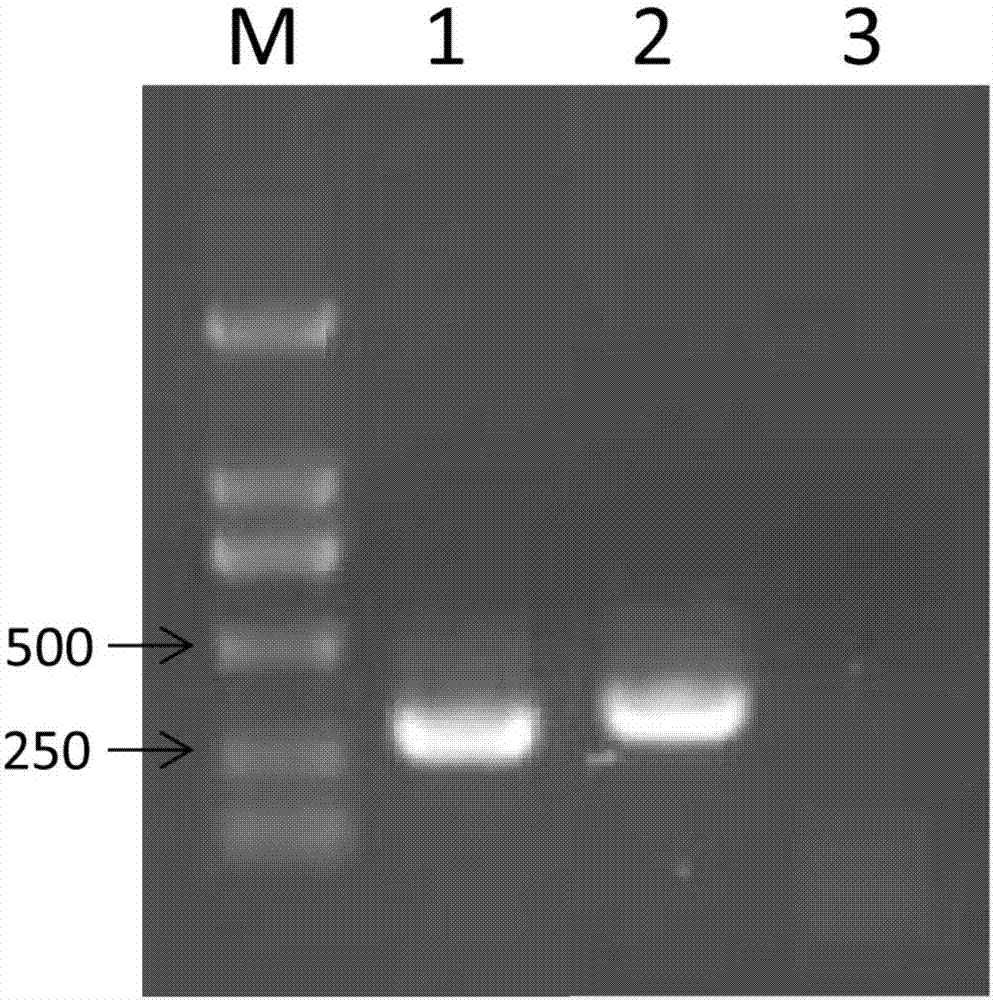
[0085] (2) PCR amplification was performed using DNA-N 23F / DNA-N 878R, and the results of electrophoresis analysis were as follows: Figure 4 shown. It can be seen from the figure that the size of the amplified fragment is about 850bp, which is consistent with the actual size;
[0086] (3) After the PCR amplification product is recovered, it is connected to the pMD18T vector;
[0087] (4) The ligation product was transformed into Esch...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com