Methods of producing a library and methods of selecting polynucleotides of interest
a polynucleotide and library technology, applied in the field of producing a library and selecting polynucleotides of interest, can solve the problems of cloning many disease genes with negative or toxic effects on cell proliferation, revertant lines are typically difficult to identify and separate from the majority of rapidly growing transformed parental cells, and each of these classical genetic approaches is limited.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction and Characterization of Vaccinia Expression Vectors
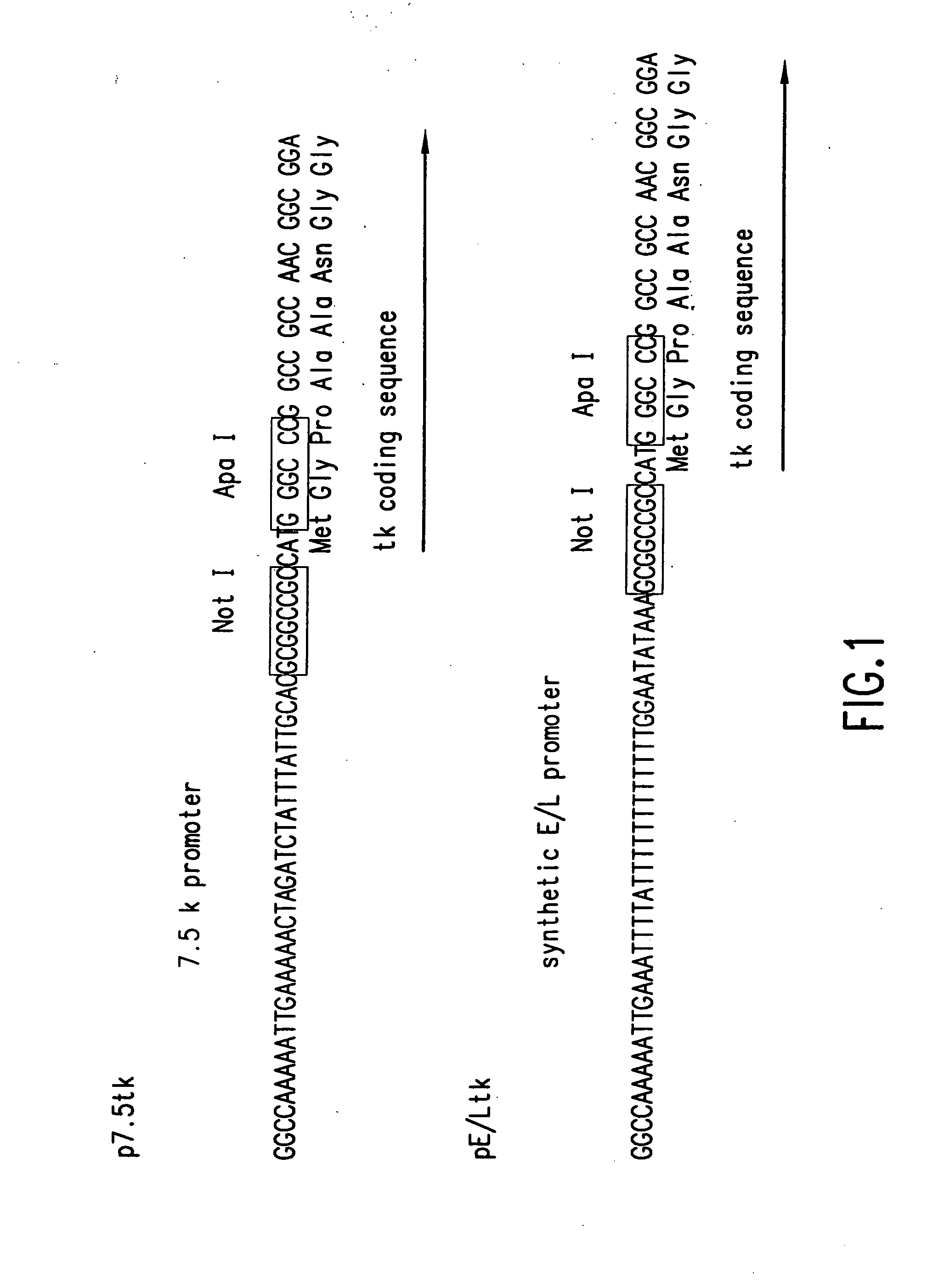
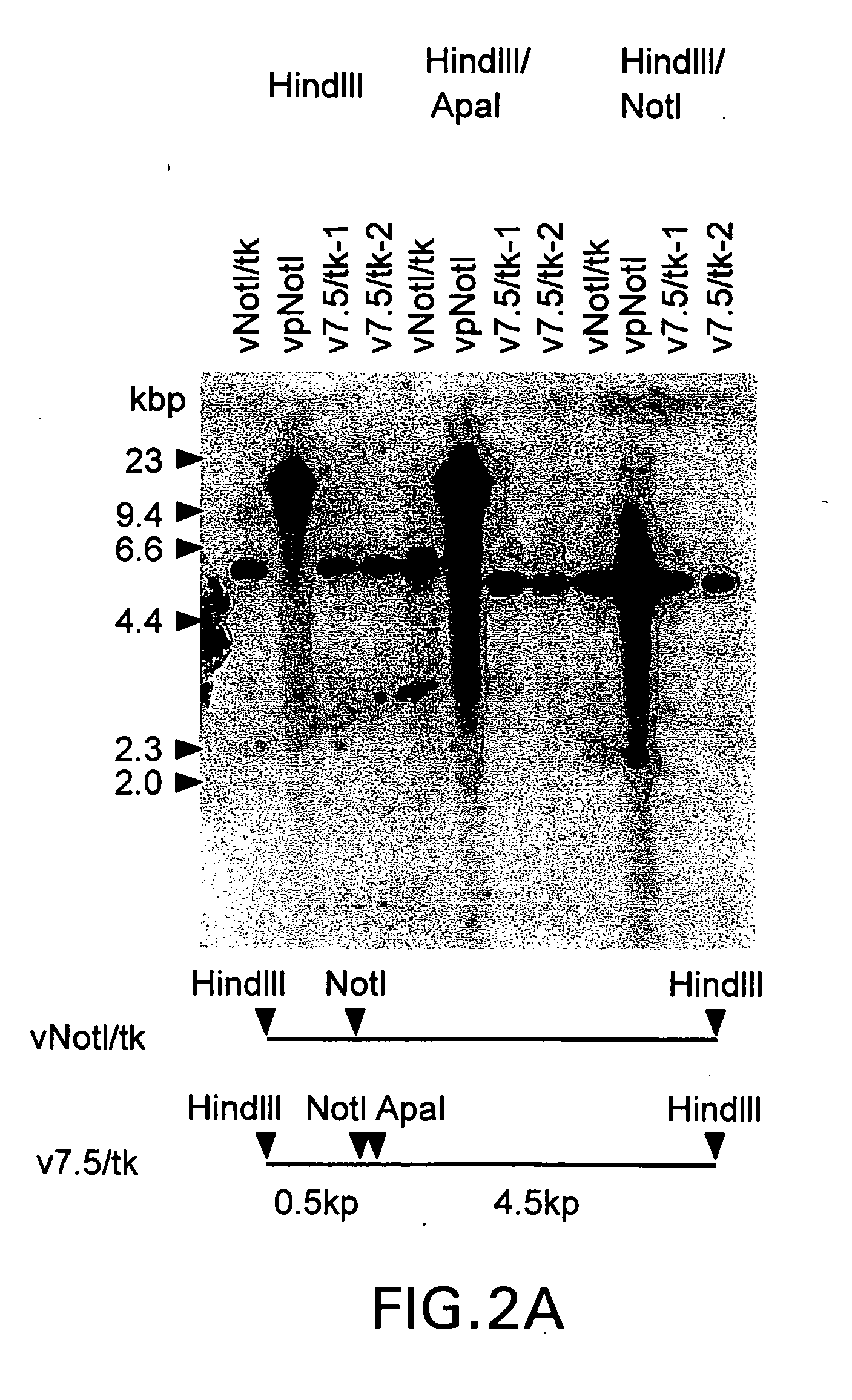
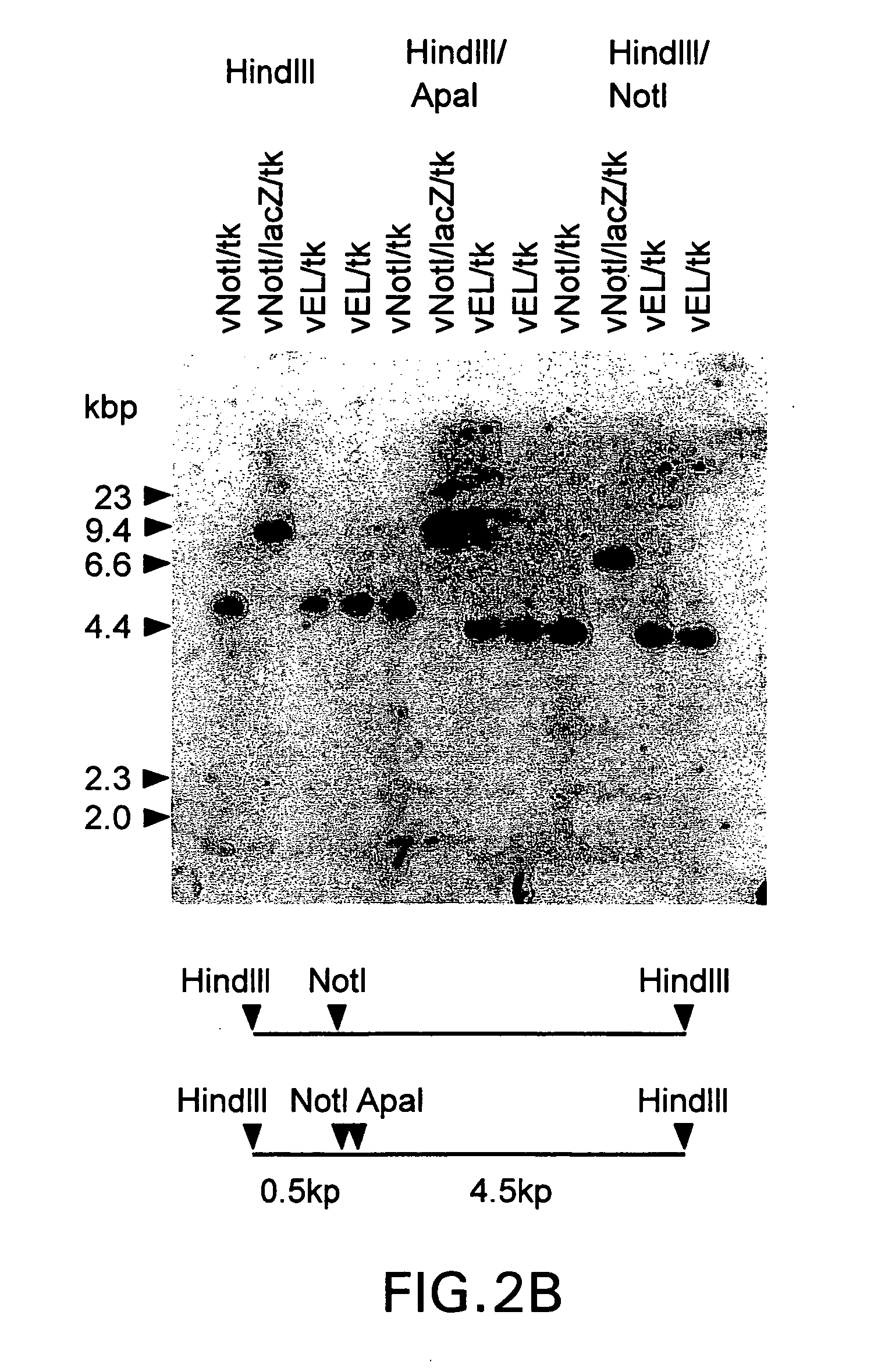
[0579] This example describes the construction and characterization of a new set of direct ligation vectors designed to be universally applicable for the generation of chimeric vaccinia genomes. The aim was to modify the genome of vNotI / tk so as to acquire direct ligation vectors which are more universally useful. First, the insertion site was changed by placing the sites for two unique restriction enzymes at the beginning of the thymidine kinase gene. This allows one to fix the orientation of the insert polynucleotide (e.g. DNA) and eliminates the production of contaminating wild type genomes after religation of viral arms. Second, in order to generate a direct ligation vector which would express high levels of protein, the thymidine kinase gene was preceded by a strong constitutive vaccinia virus promoter.
[0580] These new ligation vectors contain a pair of unique restriction sites, NotI and ApaI, to eliminate religa...
example 2
Trimolecular Recombination
[0618] Production of an Expression Library. This example describes a tri-molecular recombination method employing modified vaccinia virus vectors and related transfer plasmids that generates close to 100% recombinant vaccinia virus and, for the first time, allows efficient construction of a representative DNA library in vaccinia virus.
[0619] Construction of the Vectors. The previously described vaccinia virus transfer plasmid pJ / K, a pUC 13 derived plasmid with a vaccinia virus thymidine kinase gene containing an in-frame Not I site (Merchlinsky, M. et al., Virology 190:522-526), was further modified to incorporate a strong vaccinia virus promoter followed by Not I and Apa I restriction sites. Two different vectors, p7.5 / tk and pEL / tk, included, respectively, either the 7.5K vaccinia virus promoter or a strong synthetic early / late (E / L) promoter (FIG. 1). The Apa I site was preceded by a strong translational initiation sequence including the ATG codon. Th...
example 3
Direct Selection Using Target Epitope-Specific Cytotoxic T Cells
[0641] In this example, a model system was assayed to determine the level of enrichment that can be obtained through a procedure that selects for DNA recombinants that encode the target epitopes of tumor specific cytotoxic T cells.
Methods and Results
[0642] A specific vaccinia recombinant that encodes a well characterized ovalbumin peptide (SIINFEKL) (SEQ ID NO:26) was diluted with non-recombinant virus so that it constituted either 0.2%, 0.01%, or 0.001% of viral pfu. This ovalbumin peptide is known to be processed and presented to specific CTL in association with the murine class I MHC molecule H-2Kb. An adherent monolayer of MC57G cells that express H-2Kb were infected with this viral mix at m.o.i.=1 (approximately 5×105 cell / well). MC57G cells do not themselves express ovalbumin peptide, but do express H-2Kb, which allows them to associate with and present ovalbumin peptide to the T cells.
[0643] Following 12 hou...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com