Fluorescent quantitation PCR detection kit and detection method for PIK3CA (phosphatidylinositol3-kinase catalytic alpha) gene mutations
A detection kit and fluorescence quantitative technology, applied in the field of molecular biology, can solve the problems of limited detection accuracy, easy pollution and complicated operation of direct sequencing method, and achieve the effects of fast detection speed, strong specificity and strong selectivity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
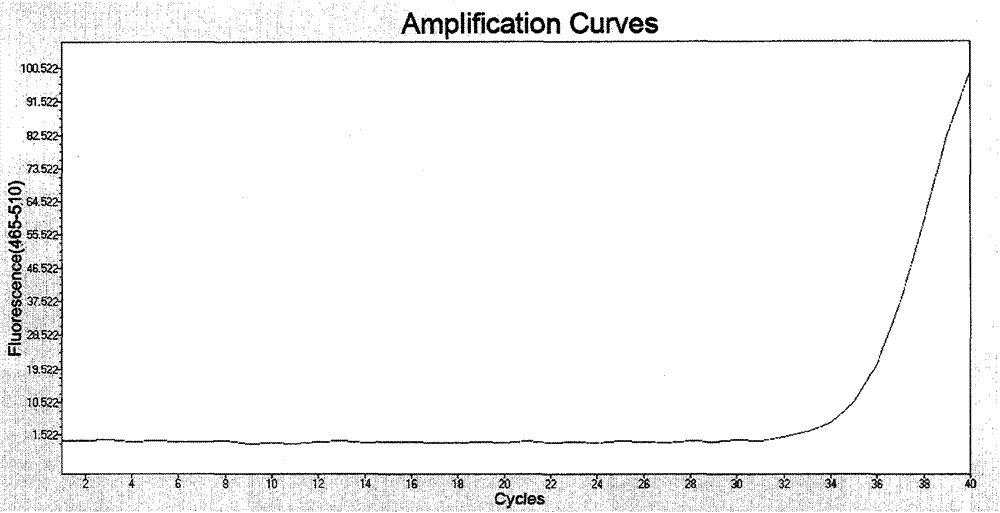
[0034]Example 1: Detection of PIK3CA gene A3140G (H1047R), A3140T (H1047L) mutants
[0035] Tumor tissues (including fresh tissues and paraffin sections) of 68 patients clinically diagnosed as colon cancer were collected, and genomic DNA was extracted as a template. Among them, about 1.0 g of fresh pathological tissue was taken, and genomic DNA was extracted using a DNA extraction kit from Qiagen; paraffin section samples were cut into sections of about 10 μm, dewaxed with xylene, and extracted using a paraffin-embedded DNA extraction kit from Qiagen Extract genomic DNA. The extracted genomic DNA was dissolved in TE buffer, that is, 10mmol / L Tris-HCl (pH8.0) and 1mmol / L EDTA (pH8.0), and the concentration was measured by an ultraviolet spectrophotometer to prepare a 50ng / μL solution. as a template for PCR amplification.
[0036] Primers and fluorescent probes capable of specifically detecting PIK3CA gene A3140G (H1047R) and A3140T (H1047L) mutations were designed, and the se...
Embodiment 2
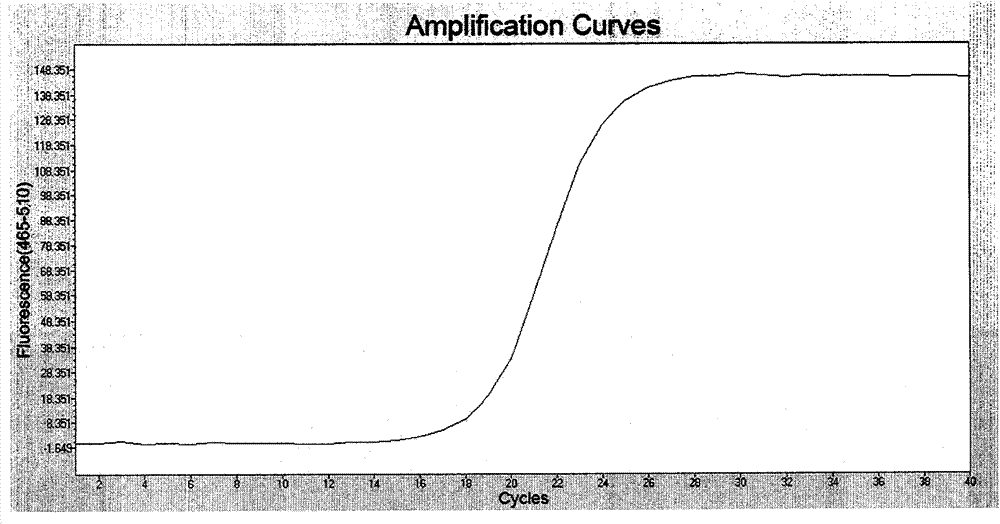
[0042] Example 2: Detection of G1633A (E545K) mutation in exon 9 of PIK3CA gene
[0043] One strain of the plasmid template containing the G1633A (E545K) mutation was used in the experiment, and two strains of cell lines were respectively H1650 and H460 (both PIK3CA gene wild type. Utilize the method for detecting PIK3CA gene No. 9 exon G1633A (E545K) mutation provided by the invention as follows:
[0044] Genomic DNA of plasmids and cell lines was extracted using Qiagen's DNA extraction kit. The extracted genomic DNA was dissolved in TE buffer, that is, 10mmol / LTris-HCl (pH8.0) 1mmol / LEDTA (pH8.0), the concentration was measured by a UV spectrophotometer, and a solution of 50ng / μL was made into a solution for PCR Amplified template.
[0045] Primers and fluorescent probes capable of specifically detecting the G1633A (E545K) mutation of the PIK3CA gene were designed, and the sequences are shown in Table 2.
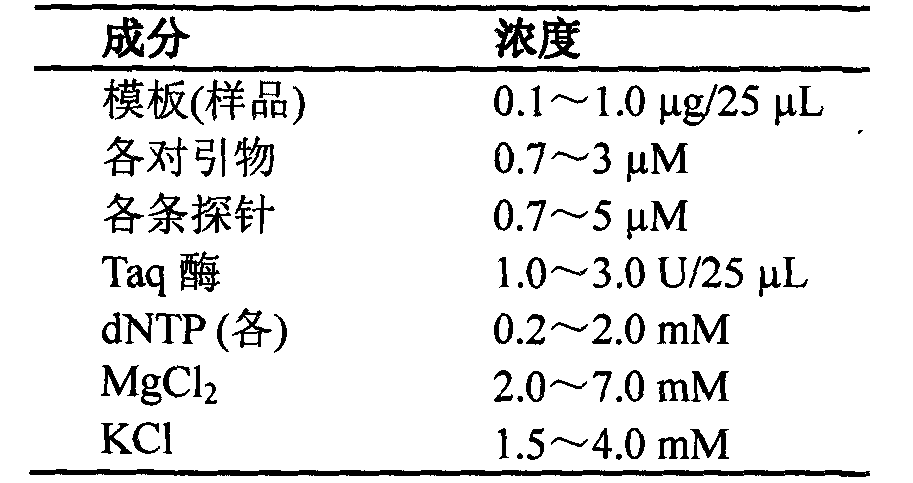
[0046] The total volume of the fluorescent PCR amplification reac...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com