Escherichia coli for producing riboflavin and constructing method and use of Escherichia coli
A technology of Escherichia coli and construction method, applied in the field of production of riboflavin, strains of Escherichia coli and construction, can solve the problems such as the production of riboflavin by Escherichia coli, and achieve the effect of improving yield and yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
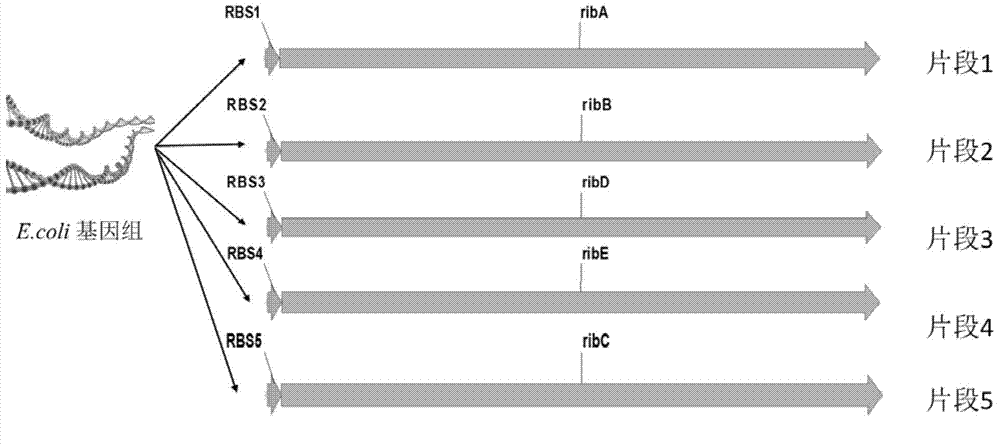
[0034] Design the ribosome binding site RBS1 shown in SEQ ID NO.37, the ribosome binding site RBS2 shown in SEQ ID NO.38, the ribosome binding site RBS3 shown in SEQ ID NO.39, and Ribosome binding site RBS4 shown in SEQ ID NO.40 and ribosome binding site RBS5 shown in SEQ ID NO.41.
Embodiment 2
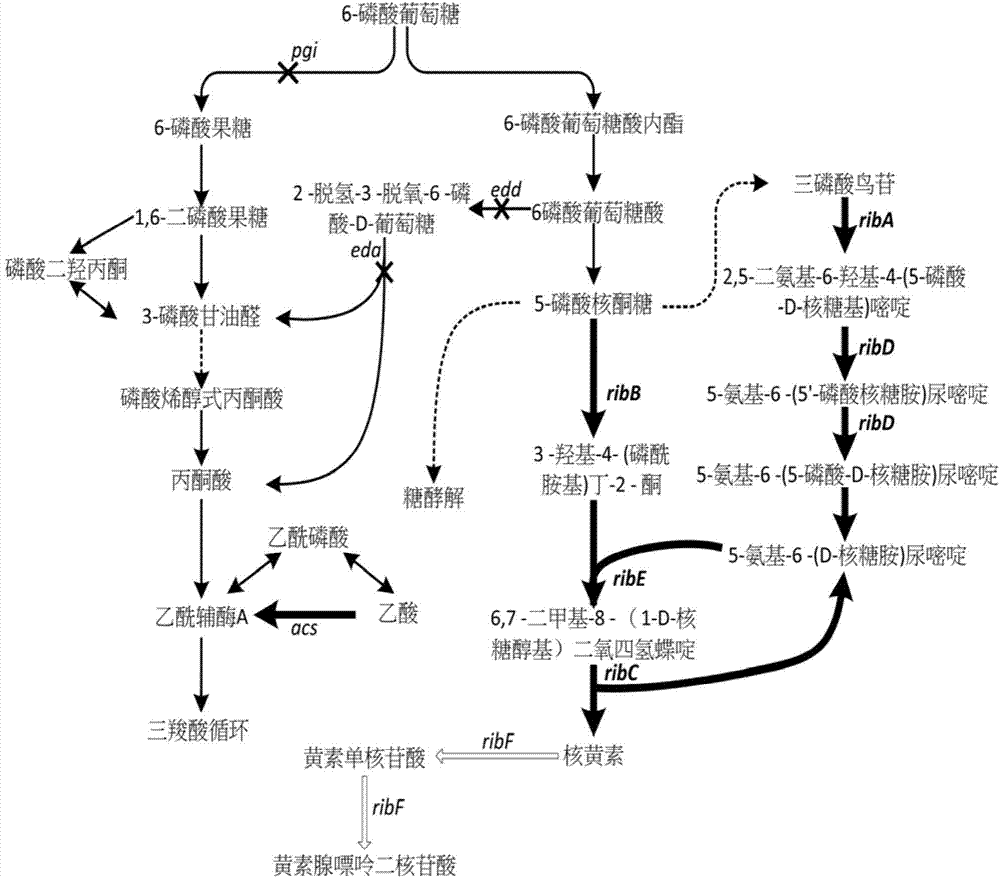
[0036] Using amplification primers FuECribA-F (SEQ ID NO.1) / FuECribA-R (SEQ ID NO.2) to use the genome of Escherichia coli MG1655 (purchased from CGSC) as a template to artificially design the ribA gene on the riboflavin synthetic metabolic pathway The ribosome binding site RBS1 and the coding sequence were amplified to obtain Fragment 1 (see figure 2 ).
[0037] Using amplification primers FuECribB-F (SEQ ID NO.3) / FuECribB-R (SEQ ID NO.4) to use the genome of Escherichia coli MG1655 (purchased from CGSC) as a template to artificially design the ribB gene on the riboflavin synthetic metabolic pathway The ribosome binding site RBS2 and the coding sequence were amplified to obtain Fragment 2 (see figure 2 ).
[0038]Using amplification primers FuECribD-F (SEQ ID NO.5) / FuECribD-R (SEQ ID NO.6) to use the genome of Escherichia coli MG1655 (purchased from CGSC) as a template to artificially design the ribD gene on the riboflavin synthetic metabolic pathway The ribosome binding...
Embodiment 3
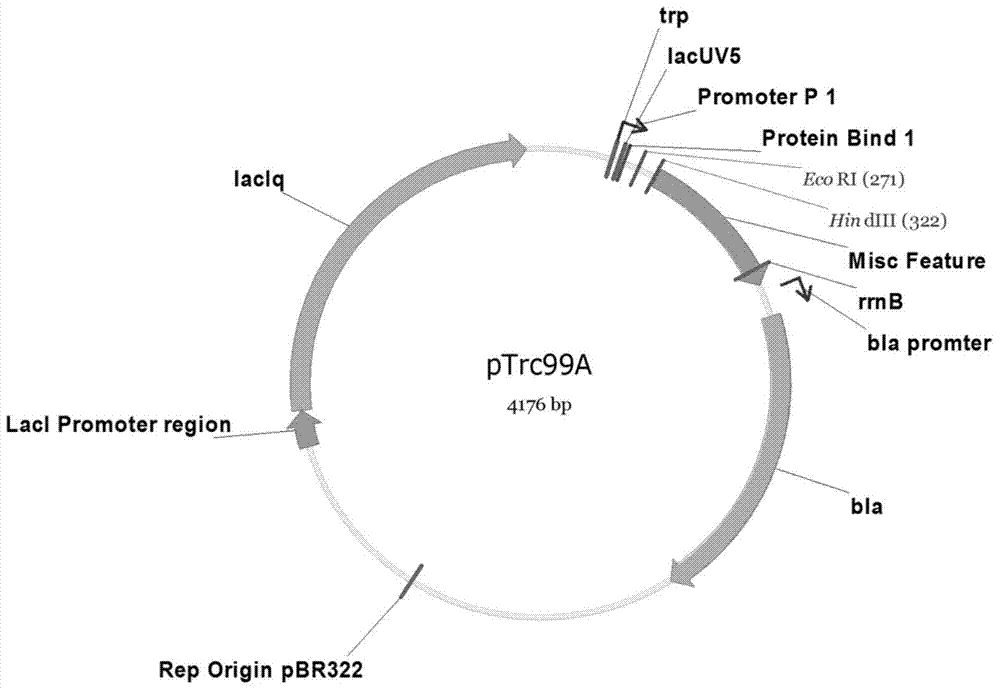
[0042] Using amplification primers FuECribA-F (SEQ ID NO.1) / FuECribC-R (SEQ ID NO.10), using fragment 1, fragment 2, fragment 3, fragment 4 and fragment 5 obtained in Example 2 as templates, by Overlap-extension polymerase chain reaction spliced these five genes in the order of RBS1-ribA-RBS2-ribB-RBS3-ribD-RBS4-ribE-RBS5-ribC and named the operon EC10 (see Figure 4 ), cloned into plasmid pTrc99a at the EcoRI / HindIII site (see image 3 , purchased from Addgene) after digestion, enzyme ligation, transformation, verification and other operations, the plasmid p20C-EC10 was obtained (see Figure 5 ), and the plasmid was transformed into Escherichia coli Escherichia coli MG1655 to obtain the RF01S strain.
[0043] Among them, the formula of LB liquid medium is: 10g / L peptone, 5g / L yeast extract, 10g / L NaCl, adjust the pH to 7.0, and sterilize under 0.1Mpa pressure for 20min. The formula of LB solid medium is: add 15g / L agar powder to LB liquid medium with adjusted pH value, an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com