GPA2 gene controlling seeds per ear of plant and applications thereof
A technology of grain number per spike and gene, applied in the field of GPA2 gene controlling the number of spikelets of plants, can solve the problems of the number of spikelets and high-level branches, low yield, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Embodiment 1, the discovery of controlling grain number per ear gene GPA2
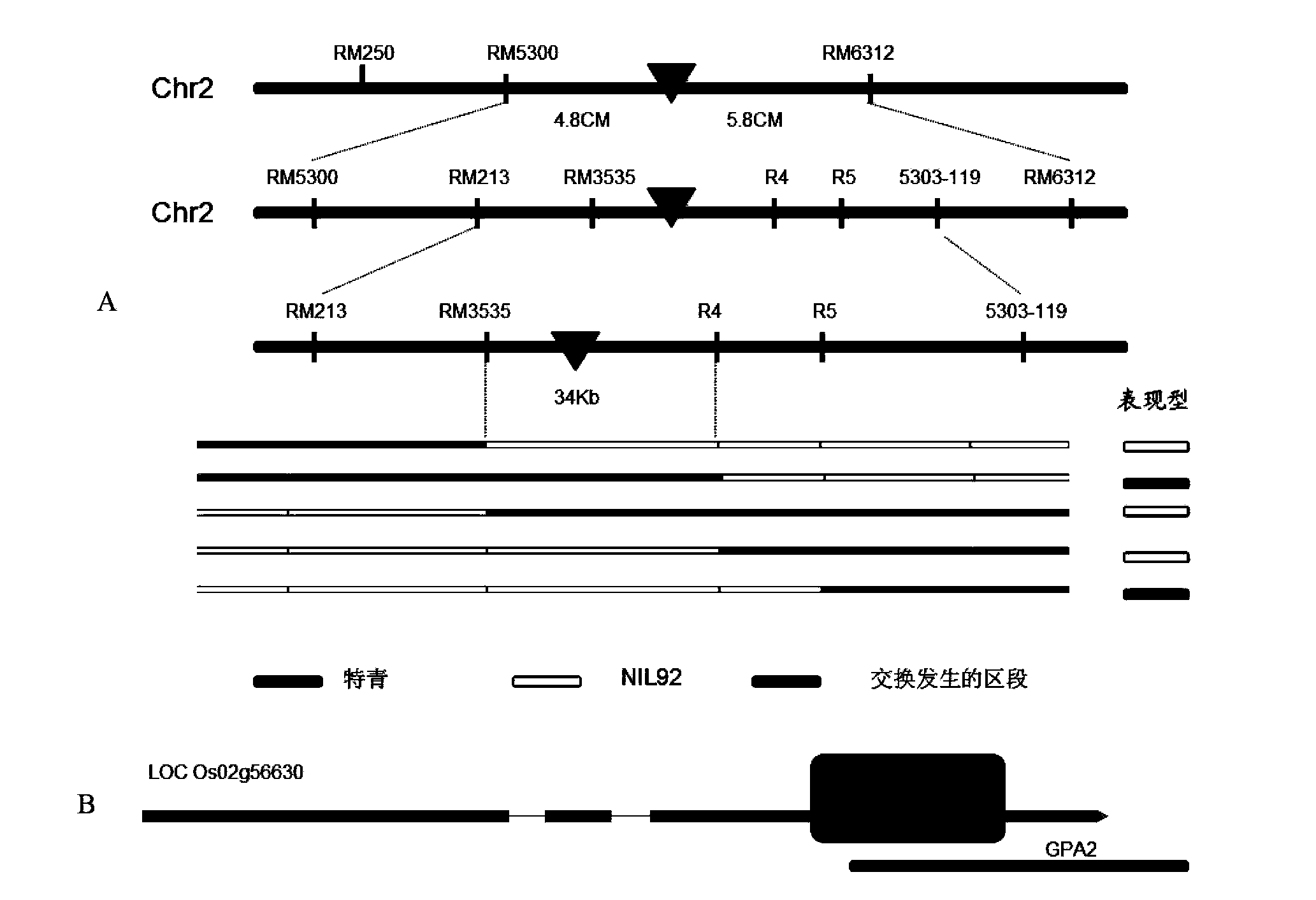
[0033] One of the introgression lines, NIL92, was found from a set of introgression lines constructed by using the common wild rice in Yuanjiang, Yunnan as the donor parent and the excellent cultivated rice variety Teqing as the recipient parent. The number of secondary branches and grains per panicle of the introgressed segment of Oryza sativa in Yuanjiang was significantly lower than that of the recurrent parent Teqing. Introduce NIL92 and Teqing ( figure 1 ) backcross and then selfcross to construct F 2 The population was identified for genotype and phenotype, and a QTL related to the number of grains per panicle was detected at the end of the long arm of the second chromosome, named q-GPA2, and the allele from NIL92 was completely dominant relative to the allele from Teqing of. The genetic linkage map of the GPA2 region was constructed using the above population, and the target QTL was pr...
Embodiment 2
[0034] Embodiment 2, the acquisition of GPA2 transgenic rice
[0035] 1. Construction of Interference Fragments
[0036] 1. Obtaining DNA Fragment A and DNA Fragment B
[0037] (1) Design primers
[0038] Recognition sites for restriction endonucleases BamHI, KpnI, SpeI and SacI were respectively introduced at both ends of the primers. The primer sequences are as follows:
[0039] CRI1-F (Forward): 5'CC GGATCC CTCCTCTGCATTGTCTTCAG-3'The underlined base is the recognition site of the restriction endonuclease BamHI;
[0040] CRI1-R (reverse primer): 5'GG GGTACC ATGGGATTTACGGTGATTGA-3'underlined base is the recognition site of restriction endonuclease KpnI;
[0041] CRI2-F:5'TG ACTAGT GCTACGCCACTGCATAAG-3'The underlined base is the recognition site of restriction endonuclease SpeI;
[0042] CRI2R:5'TA GAGCTC GTTACGGCCTTCTTCCAAAT-3'The underlined base is the restriction endonuclease SacI recognition site;
[0043](2) Obtaining DNA Fragment A and DNA Fragment B
[004...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com