Method for extracting polyribosomes from rice based on immunoprecipitation
A polyribosome and immunoprecipitation technology, applied in the field of molecular biology, can solve the problems of complex operation process, high requirements on experimental equipment and low product yield.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
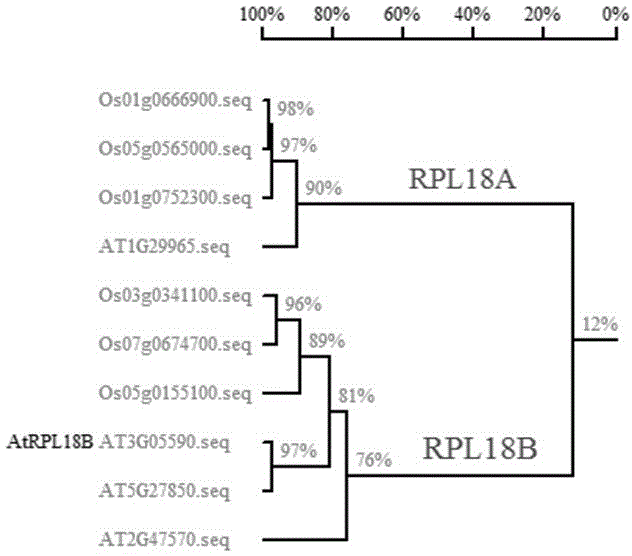
[0037] Construction of OsRPL18B1 (Os03g0341100) and OsRPL18B2 (Os05g0155100) plant expression vectors fused with Flag tags. There are 6 genes encoding RPL18 protein in rice, of which 3 belong to RPL18A and the other 3 belong to RPL18B. The present invention selects OsRPL18B1 (Os03g0341100) , OsRPL18B2 (Os05g0155100) two genes added tags, further experiments ( figure 1 ).
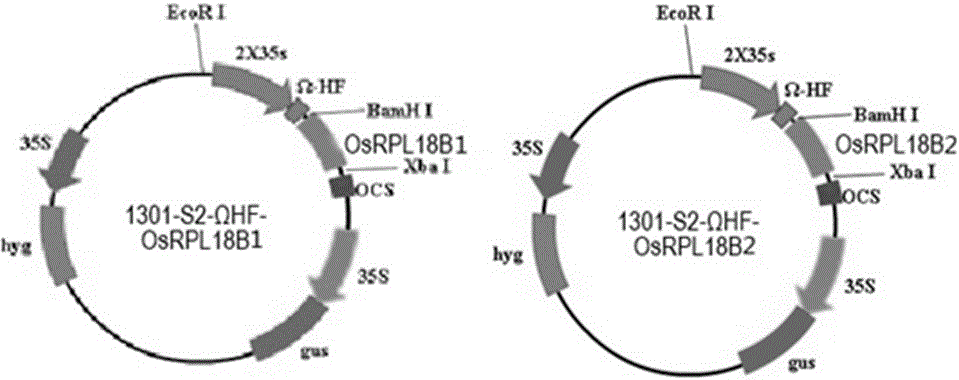
[0038] We constructed plant expression vectors of OsRPL18B1 (Os03g0341100) and OsRPL18B2 (Os05g0155100) fused with Flag tags, and transformed them into rice Nipponbare respectively, so that the fusion proteins were overexpressed under the promoter of 35S promoter and Ω-HF enhancer ( figure 2 ).
Embodiment 2
[0039]Molecular identification of embodiment 2 transgenic strains
[0040] Since we need to obtain as many tagged ribosome-mRNA complexes as possible in the transgenic lines, GUS staining and western blot identification were performed on the obtained transgenic lines, and the lines with higher expression of fusion protein were selected for further analysis. further research.
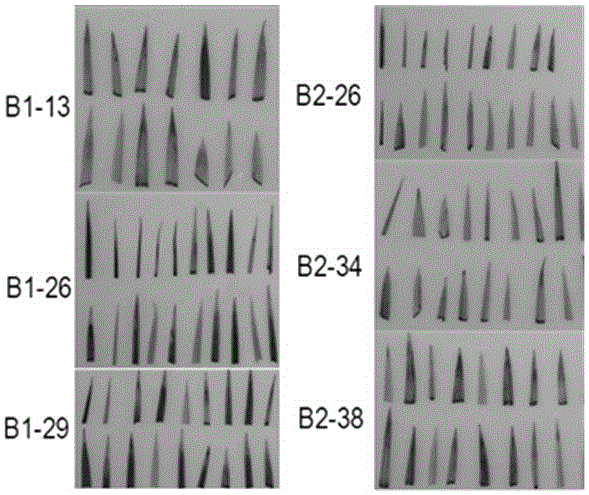
[0041] Stained by GUS ( image 3 ) and western blot ( Figure 4 ) identification, we selected four strains B1-13, B1-29, B2-26, B2-38 for the next experiment.
Embodiment 3
[0042] The extraction of embodiment 3 polyribosomes
[0043] Ribosomes can remain bound to mRNA in the presence of high magnesium ions, so EGTA is used to chelate other metal ions in tissue homogenate; adding chloramphenicol and cycloheximide can also inhibit the dissociation of ribosomes and mRNA .
[0044] All experimental equipment must be free from RNase contamination. According to different materials, RNase can be removed by drying at 160°C for 4 hours, soaking in 0.1% DEPC or 10% hydrogen peroxide and autoclaving. Non-alcoholic solutions need to be prepared with RNase free milliQ water. In the experiment, try to keep the operation on ice or at 4°C.
[0045] 1) Fragmentation of plant tissue
[0046] In the following experiments, 1% volume of 0.1M PMSF was added to the current system every half hour;
[0047] Add twice the volume of PEB to the liquid nitrogen ground material powder and co-grind with liquid nitrogen (at least 2ml sample). The glass homogenizer in the o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com