Digital PCR platform based gene deletion mutation detection method and kit thereof
A gene deletion and kit technology, applied in the field of molecular biology, can solve problems such as poor sensitivity and inability to meet high-sensitivity detection requirements, and achieve the effect of improving detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
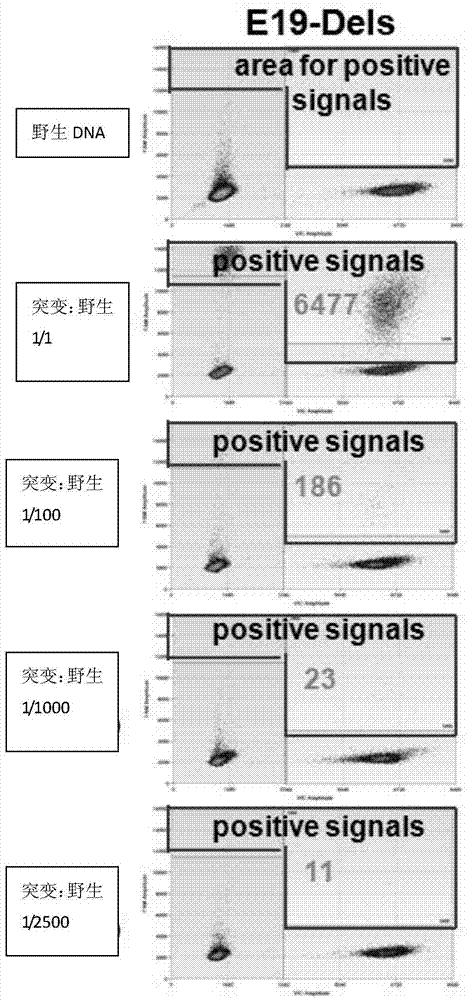
[0044] (1) Prepare DNA samples to be tested: DNA containing wild-type EGFR gene, EGFR exon 19 deletion mutation-positive DNA mixed with wild-type DNA (the ratio of mutant to wild-type content is 1 / 1, 1 / 100, 1 / 1000, 1 / 2500). The source of DNA can be serum, plasma, peripheral blood, oral mucosa, pleural effusion, body fluid or tissue, etc. The mutant DNA was derived from a positive cell line carrying the EGFR19 mutation. Among them, the c.2238-c.2255 deletion mutation on the EGFR19 exon.
[0045] (2) Melt forward and reverse detection PCR amplification primers and corresponding labeled TaqMan probes and PNA probes for detecting EGFR exon 19 deletion at room temperature, as well as forward and reverse quality control PCR primers and quality control PCR primers. control TaqMan probes.
[0046] The sequences of forward and reverse PCR amplification primers are:
[0047] F1: 5'-GAGAAAGTTAAAATTCCCGTCGCT-3'
[0048] R1:5'-ATGGACCCCCCACACAGCA-3'
[0049] The sequence of the label...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com