Circulating tumor cell identification kit and circulating tumor cell identification method
A tumor cell and identification method technology, which is applied in the field of medical and biological, circulating tumor cell identification kits, can solve the problems of high requirements, contaminated blood samples, false positives, etc., and achieve the goal of improving sensitivity, detection sensitivity, and fluorescence signal intensity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] There are two types of circulating tumor cell identification kits described in this example, those with labeled probes and those without labeled probes.
[0048] Among them, the circulating tumor cell identification kit A with labeled probes mainly includes:
[0049] 1. Capture probe
[0050] The capture probe is composed of three parts, the 5' end to the 3' sequence is the sequence P1 complementary to the mRNA of the corresponding marker gene, the spacer sequence, and the P2 sequence complementary to the corresponding amplification probe specific sequence P3, Marker genes of the same category have the same P2 sequence in their capture probes. The spacer is used to space the capture probe P2 sequence from the target mRNA, and by setting a spacer sequence of appropriate length inside the probe, it can reduce steric hindrance, improve the efficiency of the hybridization reaction and the specificity of the hybridization reaction . The spacer arm of the capture probe of ...
Embodiment 2
[0077] Example 2 Using the kit A in Example 1 to detect circulating tumor cells in human peripheral blood
[0078] The formula of described various solutions is as follows:
[0079]
[0080]
[0081] All the probes in the corresponding gene list in Example 1 were used in the probe mixture, amplification mixture, and chromogenic mixture in this example.
[0082] 1. Sample pretreatment, filter CTCs onto the filter membrane
[0083] 1. Preserve the blood sample in RI preservation solution in the sample preservation tube, centrifuge at 600×g for 5 minutes, discard the supernatant, and remove the red blood cells.
[0084] 2. Add 4mL PBS and 1mL RI fixative, vortex and mix well, and let stand at room temperature for 8min.
[0085] 3. Sample filtration: Transfer the liquid in the sample storage tube to the filter, turn on the vacuum filtration pump to pump out the liquid; add 4mL PBS to the storage tube, wash the tube wall and filter the liquid.
[0086] 4. Transfer the filt...
Embodiment 3
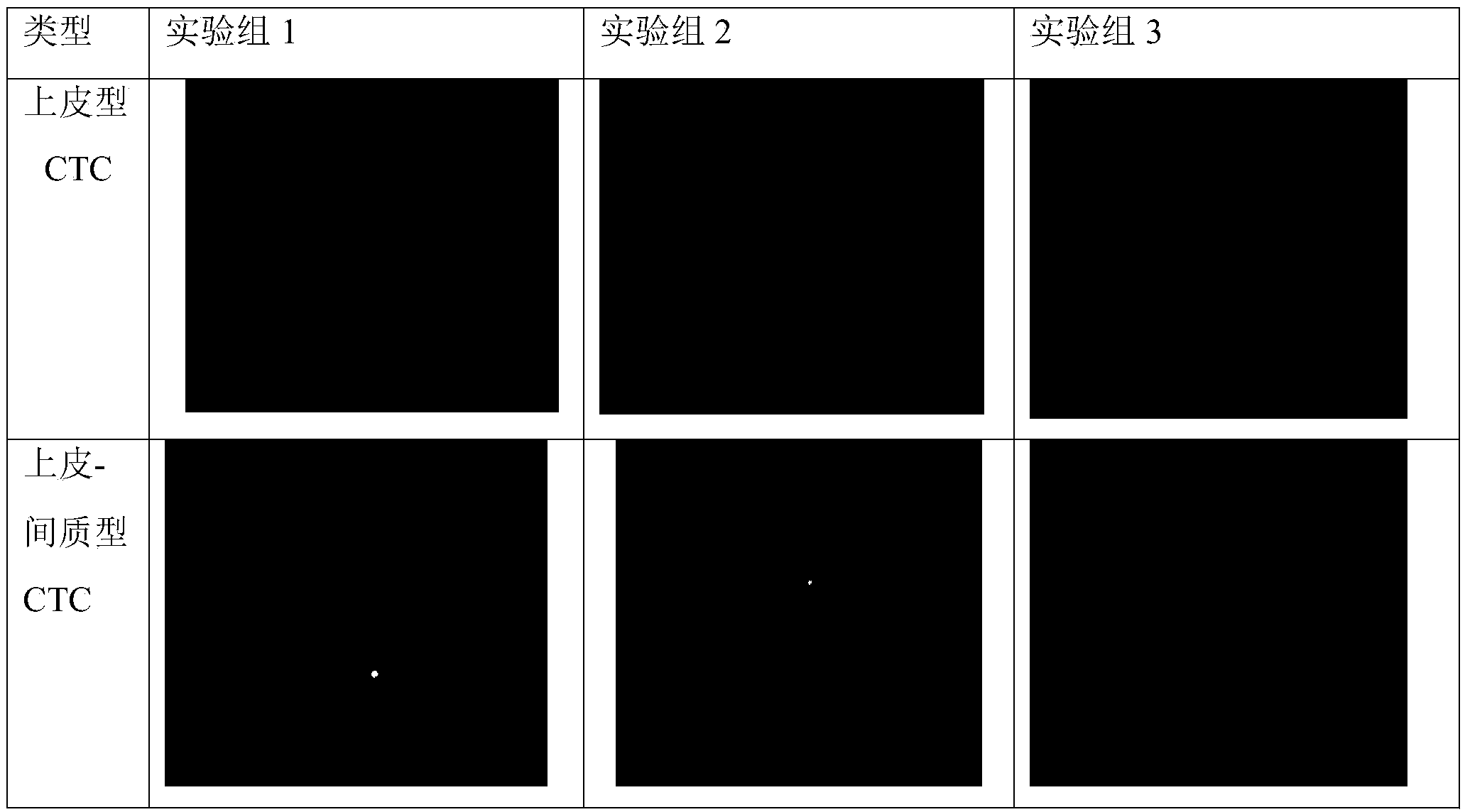
[0146] Example 3 Selection of the target detection marker gene of the kit
[0147] 1. Design of kit preparation (selection of the number and types of target detection marker genes)
[0148] The epithelial cell marker gene of the kit of the present invention is selected from: EPCAM, KRT16, KRT8, KRT18, KRT19, and the mesenchymal cell marker gene is selected from: CDH2, VIMENTIN, FN1, AKT2. When the epithelial cell marker gene and the mesenchymal cell marker gene are selected differently When the number and type of genes were tested, the detection results were consistent.
[0149] The selection of epithelial cell marker genes as an example, refer to the implementation group 1-3, select one, three and five marker genes respectively, and compare their detection results, while mesenchymal cell marker genes use all 4 genes and The specific design of leukocyte marker gene CD45 is shown in Table 10.
[0150]Taking leukocyte marker genes as an example, experimental group 4 used all e...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Membrane pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com