Construction and application of Zymomonas mobilis CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-association proteins)9 system
A Zymomonas, psuzm1a-cas9 technology, applied in the construction and application of Zymomonas mobilis CRISPR-Cas9 system, can solve the problems of complex experimental process and low efficiency, and achieve the effect of good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
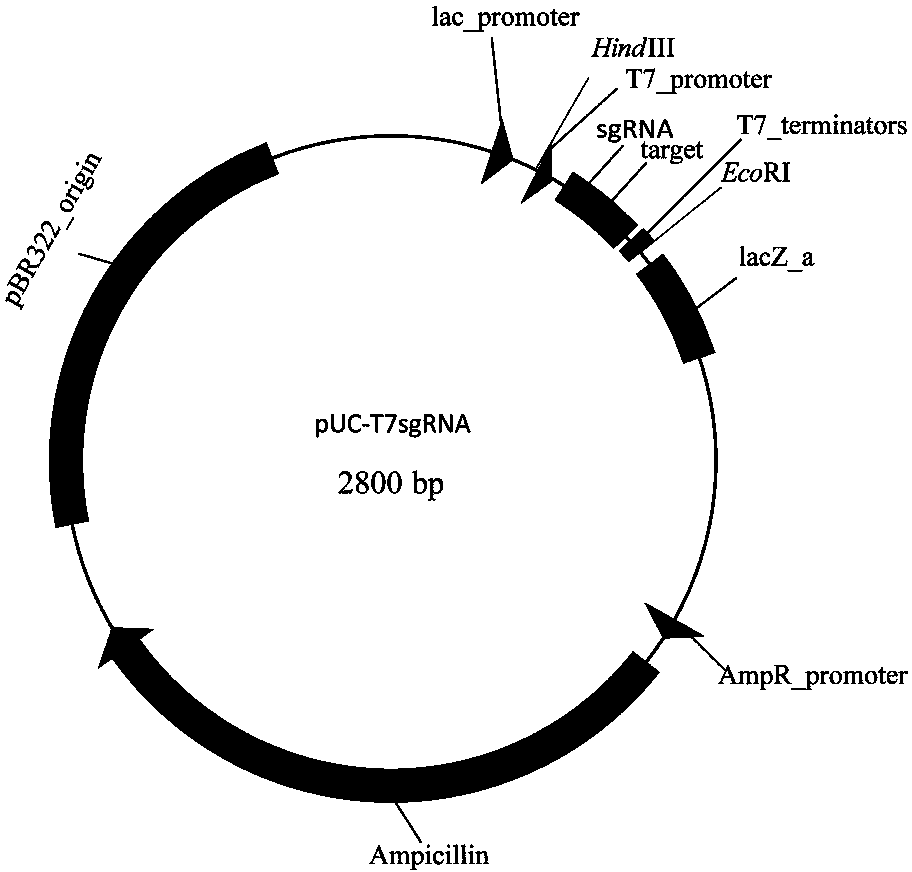
[0045] Example 1 Construction of expression plasmid pUC-T7sgRNA
[0046] 1) Design of T7sgRNA gene: Artificially synthesize a double-stranded DNA with a full length of 177bp, including T7 gene promoter and terminator, Bbs I recognition sequence and crRNA-tracrRNA sequence, the two ends of the sequence are respectively Hin dIII and Eco RI site.
[0047] The T7sgRNA gene sequence is as follows:
[0048] GG AAGCTT AA TACGACTCAC TATAGGTCTT CGA GAAGAC C TGTTTTAGAG CTAGAAATAG CAAGTTAAAA TAAGGCTAGT CCGTTATCAA CTTGAAAAAG TGGCACCGAG TCGGTGCTTT TTCTAGCATA ACCCCTTGGG GCCCTTAAAC GGGTCTTGAG GGGTTTTTT G AATTC CC
[0049] 2) Plasmid pUC19 and T7sgRNA gene fragments were used respectively Hin dIII and Eco RI for double digestion
[0050] 3) The digested product was ligated with T4 DNA ligase, transformed into Escherichia coli, and the expression plasmid pUC-T7sgRNA was obtained (see figure 1 ).
example 2
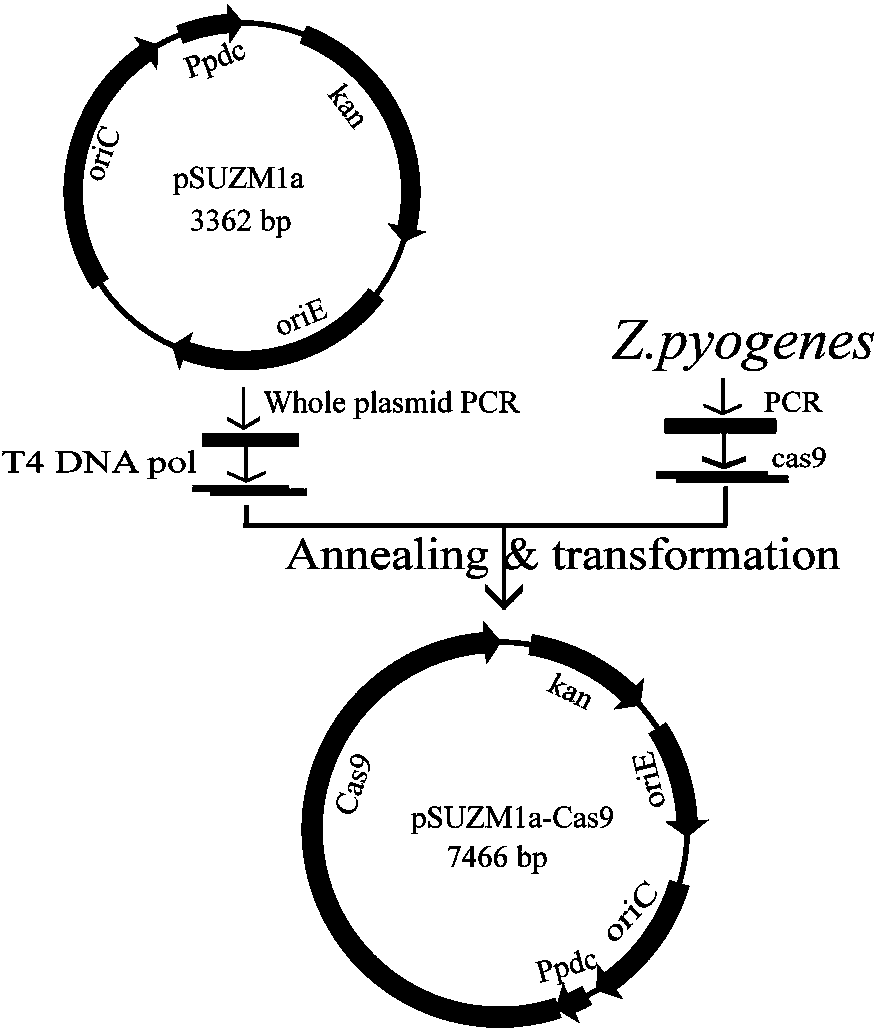
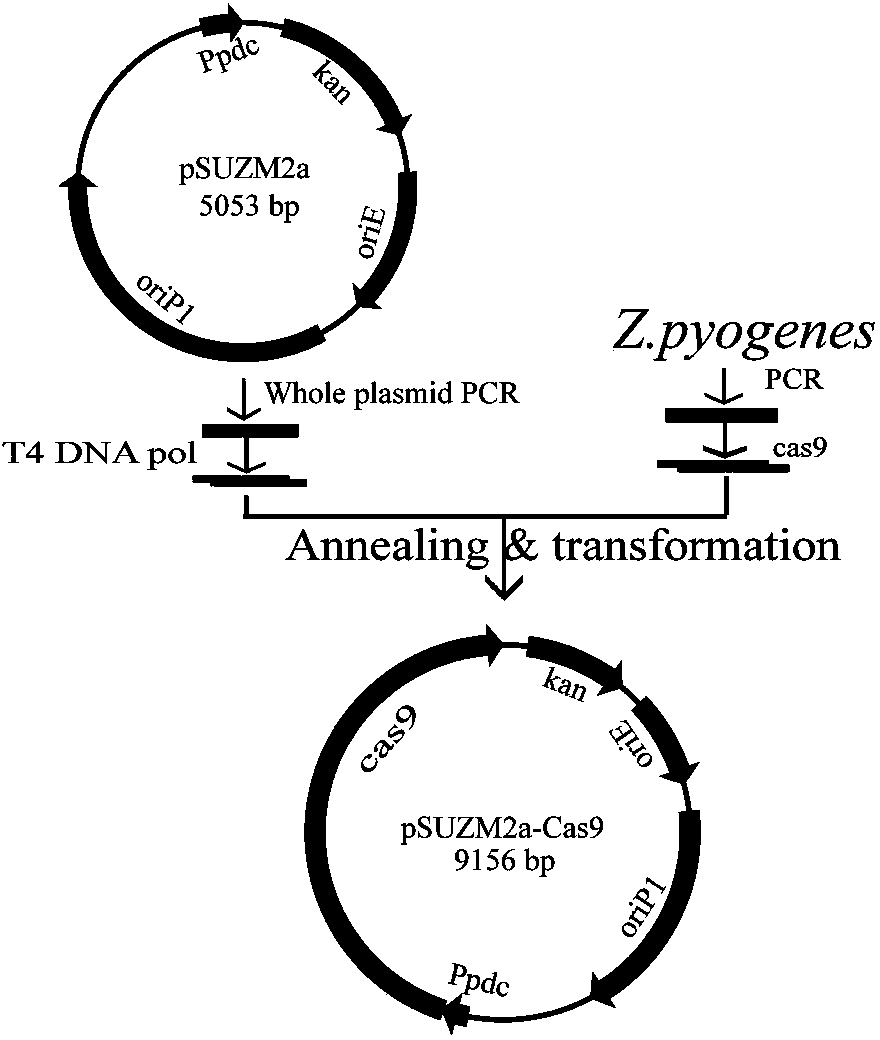
[0051] Construction of example 2 expression plasmids pSUZM1a-Cas9, pSUZM2a-Cas9, pSUZM3a-Cas9
[0052] For the construction strategy of expression plasmids pSUZM1a-Cas9, pSUZM2a-Cas9, pSUZM3a-Cas9, see figure 2 , image 3 , Figure 4 .
[0053] 1) Design primers 1V-Cas9 upstream, 1V-Cas9 downstream; primer Cas9-1 upstream, primer Cas9-1 downstream; the specific sequence is as follows:
[0054] 1V-Cas9 upstream: 5'-CTAGGAGGTGACTGAAGGTAGCTTGCAGTGGG-3'
[0055] 1V-Cas9 downstream: 5'-GAGTATTTCTATCCATTGCTTACTCCATATAT-3'
[0056] Cas9-1 upstream: 5'-ATATATGGAGTAAGCA ATGGATAAGAAATACTC-3'
[0057] Downstream of Cas9-1: 5'-CCCACTGCAAGCTACCT TCAGTCACCTCCTAG-3'
[0058] 2) Using plasmids pSUZM1a, pSUZM2a, and pSUZM3a as templates, use primers 1V-Cas9 upstream and 1V-Cas9 downstream to amplify vector backbone fragments A, B, and C; use Streptococcus pyogenes CICC10464 as a template; use primer Cas9-1 upstream and downstream of Cas9-1 to amplify the Cas9 gene fragment.
[0059] P...
example 3
[0076] Example 3 Application of CRISPR-Cas9 system in Escherichia coli
[0077] 1) Transform Escherichia coli DH5α with plasmids pSUZM1a-Cas9, pSUZM2a-Cas9, pSUZM3a-Cas9
[0078] Preparation of competent cells: Pick a single colony of Escherichia coli strain DH5α that was cultured overnight, inoculate it in 2 mL of SOB, and inoculate it with shaking at 37 °C overnight; take 0.5 mL of the bacterial liquid, inoculate it in 50 mL of SOB, and inoculate it at 18 °C for about 24 h or shake culture at 25°C for about 12 h; transfer the bacterial solution into a 50 mL centrifuge tube, bathe in ice water for 10 min, centrifuge at 4,000 rpm for 10 min, discard the supernatant; add 16 mL of 0°C pre-cooled TB buffer ( 20 mmol / L KCl, 54 mmol / L MnCl 2 , 15 mmol / L CaCl 2 , 12.5 mmol / L K-MES pH 6.2), resuspend the cells, centrifuge at 4,000 rpm for 10 min, then add 4 mL of TB buffer to suspend the cells, add 280 μL DMSO dropwise, mix well, and place in an ice bath for 10 minutes min or long...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com