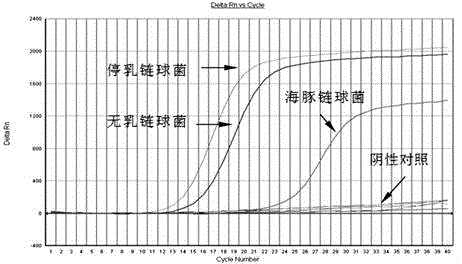
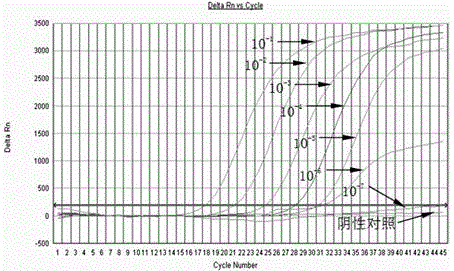
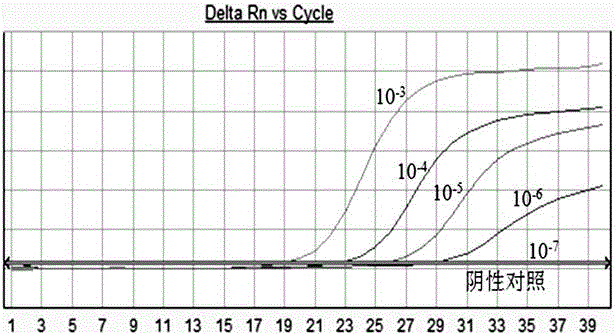
Triple real-time fluorescent PCR method and kit for detecting three streptococci at the same time
A real-time fluorescence, streptococcus technology, used in biochemical equipment and methods, recombinant DNA technology, microbial assay/inspection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] Preparation of Three Streptococcus DNA Templates
[0084] Three kinds of streptococcus [Streptococcus agalactiae (ATCC 13813), Streptococcus dysgalactiae (ATCC 35666), Streptococcus iniae (ATCC 29178) were tested according to the instructions of TIANamp Bacteria DNA Kit, and the three kinds of streptococci were obtained from the American Type Culture Collection ( ATCC)] Genomic DNA extraction and purification, add the corresponding reagents in order:
[0085] (1) Take 1.5 mL of bacterial liquid, centrifuge at 10,000 r / min for 1 min, discard the supernatant, add 200 μL of buffer GA, and suspend the bacterial cells thoroughly;
[0086] (2) Add 20 μL proteinase K (20mg / mL), and add 220 μL buffer solution GB, mix thoroughly, and act in a 70°C water bath for 10 minutes;
[0087] (3) Add 220 μL of absolute ethanol, mix thoroughly for 15 seconds, transfer the resulting solution (including flocculent precipitate) to the adsorption column CB3 after brief centrifugation, centrif...
Embodiment 2
[0094] Preparation of positive control substance
[0095] 1. Design and synthesis of PCR primers
[0096] Select the cpsF gene of Streptococcus agalactiae as the target gene, the MIG gene of Streptococcus dysgalactiae as the target gene, and the 16SrRNA gene of Streptococcus iniae as the target gene, and after BLAST analysis, design and synthesize primer pairs, as shown in Table 1:
[0097] Table 1 Three PCR primer pairs for Streptococcus
[0098]
[0099] 2. Utilize the target gene amplification primers SA-F / SA-R, SD-F / SD-R, SI-F / SI-R of the above-mentioned three kinds of streptococcus, respectively amplify Streptococcus agalactiae by PCR method cpsF gene, Streptococcus dysgalactiae MIG gene and Streptococcus iniae 16S rRNA gene. The reaction system of the three-segment gene PCR is as follows:
[0100] 10×PCR Buffer 2.5μL dNTP (2.5mM for each) 2.0 μL upstream primer 1.0 μL downstream primer 1.0 μL Taq DNA Polymerase (5U / μL) 0...
Embodiment 3
[0115] Design and synthesis of primers and probes for triple real-time fluorescent PCR of three kinds of streptococci
[0116] According to the genome DNA sequences of Streptococcus agalactiae, Streptococcus dysgalactiae and Streptococcus iniae released in Genbank, combined with bioinformatics methods, the cpsF gene of Streptococcus agalactiae, the MIG gene of Streptococcus dysgalactiae and Streptococcus iniae The conserved region sequence of the 16S rRNA gene was used as the detection target sequence, and 3 pairs of primers and 3 probes were designed and synthesized for the qualitative detection of Streptococcus agalactiae, Streptococcus dysgalactiae and Streptococcus iniae. As shown in table 2.
[0117] Table 2 Triple real-time fluorescent PCR primers and probes for three kinds of streptococci
[0118]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com