Strain capable of improving performance of recombinant protein under aerobic condition
A protein and strain technology, applied in the field of Escherichia coli strains, can solve the problems of inability to generate energy, the expression of recombinant proteins cannot meet the needs of the industry, and the expression of recombinant proteins is limited.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
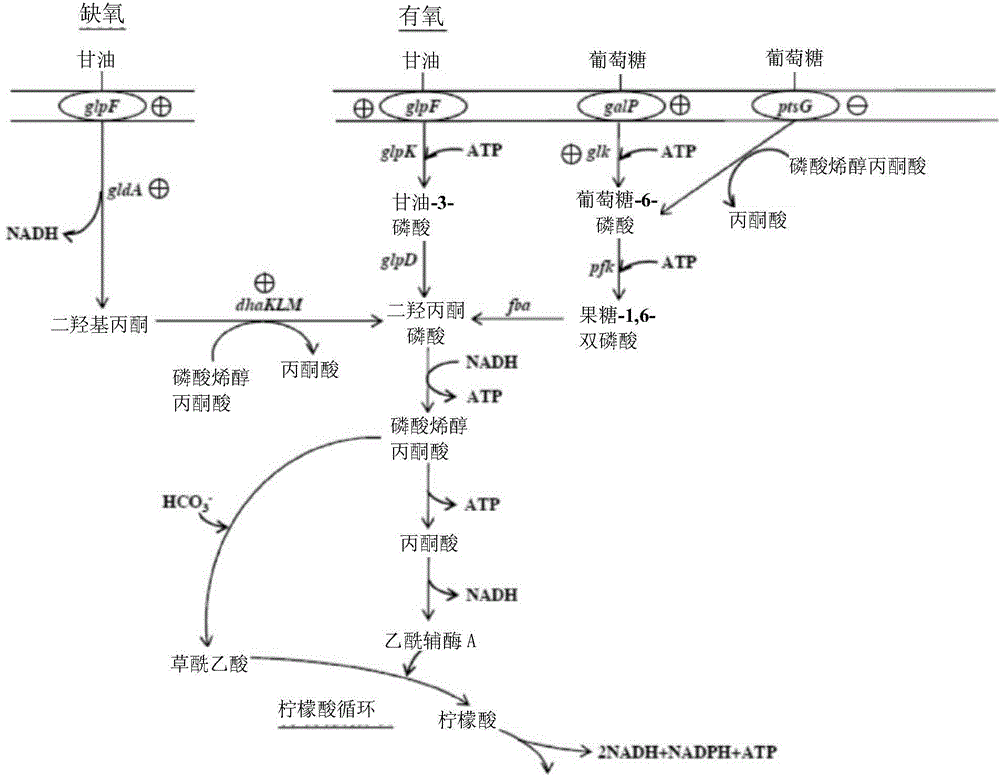
[0040] Ⅰ. Trc promoter replaces the promoter of glpF gene
[0041] A Ta1 primer (SEQ ID NO: 1) and a Ta2 primer (SEQ ID NO: 2) were designed according to the pTrc99A plasmid, and the Ta1 primer has a cutting site for the restriction enzyme EcoRI. The plasmid pTrc99A was used as a template, and the two primers were used for PCR to obtain a DNA fragment containing a Trc promoter. Design the PK1 primer (SEQ ID NO: 3) and the PK2 primer (SEQ ID NO: 4) according to the plasmid pLoxKm-PR (refer to China Taiwan Invention Patent Application No. 102144504). The PK1 primer has a cutting site for the restriction enzyme SmaI, and the PK2 primer There is a cleavage site for the restriction enzyme EcoRI. The plasmid pLoxKm-PR was used as a template, and the two primers were used for PCR to obtain a DNA fragment containing LoxP site, kanamycin resistance gene, and LoxP site sequentially. Use the restriction enzyme EcoRI to cut the former DNA fragment, and use the restriction enzyme SmaI an...
Embodiment 2
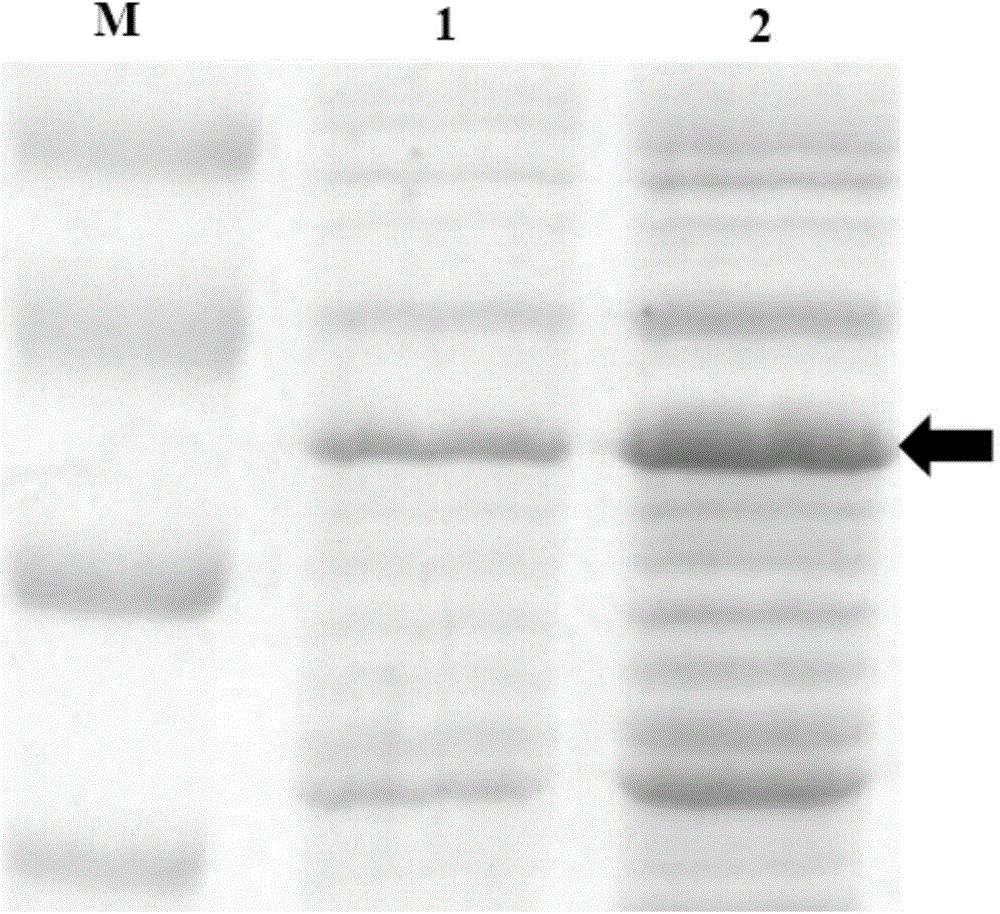
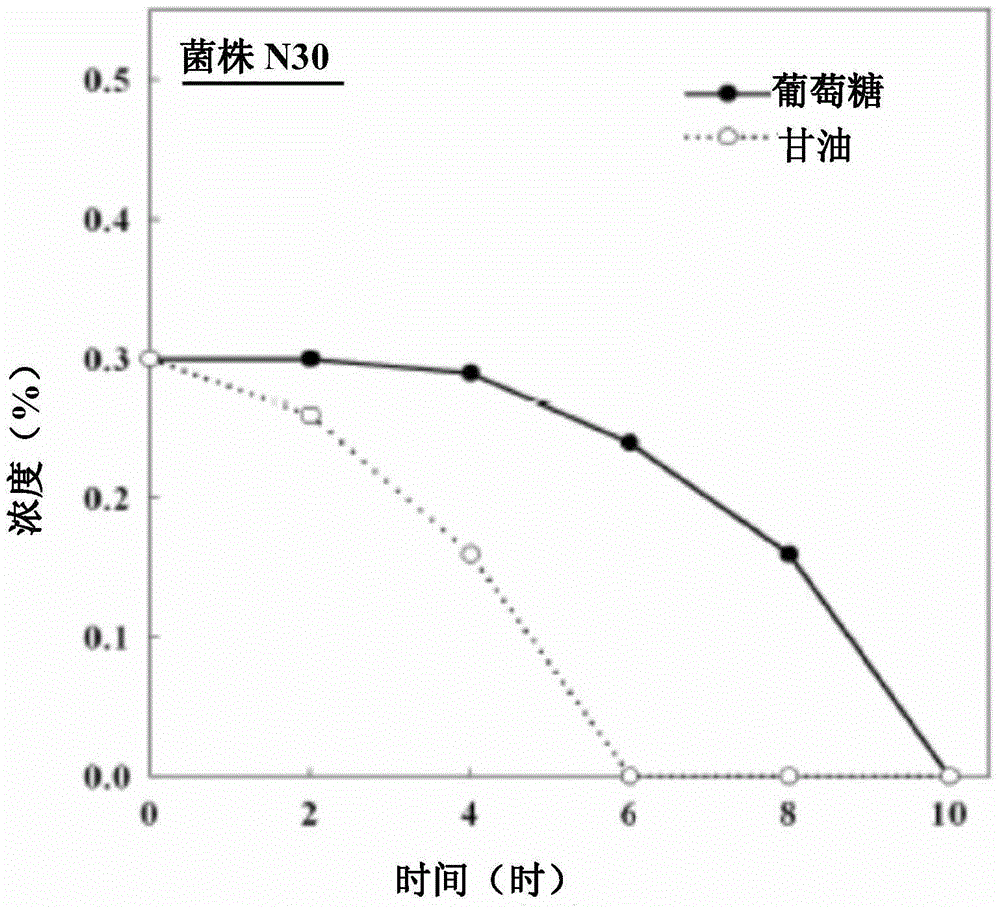
[0055] The plasmid pChHDT (refer to J Biotechnol.2005; 117(3):267-75) was transformed into strain N30 and strain BAD5 by the above-mentioned "chemical transformation method" to obtain recombinant strain N30 / pChHDT and recombinant strain BAD5 / pChHDT, wherein the plastid pChHDT contains at least a D-type hydantoinase gene operably linked to a T7 promoter.
[0056] After picking a colony of the two recombinant strains from LB solid medium to 5 mL of LB liquid medium containing 50 μg / mL ampicillin, the bacterial liquid was shaken at 200 rpm in a 30°C incubator for 16 hours, and then inoculated to 20 mL M9 liquid medium (6g / L Na2HPO4, 3g / L KH2PO4, 0.5g / L NaCl, 1g / L NH4Cl, 1mM MgSO4.7H2O, 0.1mM CaCl2), the initial concentration is OD550=0.08. Next, the bacterial solution was cultured in a 30° C. incubator with shaking at 250 rpm until its concentration reached OD550 = 0.3, and then 30 μM arabinose was added to continue culturing for 6 hours to induce the expression of D-type hydan...
Embodiment 3
[0063] Ⅰ. Trc promoter replaces the promoter of galP gene
[0064] A gal1 primer (SEQ ID NO: 22) and a gal2 primer (SEQ ID NO: 23) were designed according to the chromosome of the Escherichia coli strain BL21, the gal1 primer has a cleavage site for the restriction enzyme KpnI, and the gal2 primer has a cleavage site for the restriction enzyme SacI. Using the chromosome of the bacterial strain BL21 as a template, the two primers are used to carry out PCR on it to amplify a DNA fragment containing part of the upstream region of the galP gene and a part of the region of the galP gene. After cutting the DNA fragment and the plasmid pBlue-glpF-lox with the restriction enzyme KpnI and the restriction enzyme SacI, the cut DNA fragment and the cut plasmid were ligated with T4 cohesive enzyme to obtain the plasmid pBlue-galP, which contains A partial upstream region of the galP gene and a partial region of the galP gene.
[0065]The gal3 primer (SEQ ID NO: 24) and the gal4 primer (SE...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com