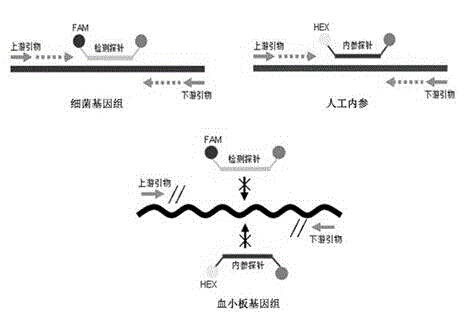
Universal type fluorescent quantitative PCR method for detecting platelet bacterial pollution
A fluorescence quantitative and detection method technology, applied in the direction of biochemical equipment and methods, microbial measurement/testing, etc., can solve the problems of complex result interpretation standards, poor sensitivity, long detection time, etc., to achieve effective auxiliary diagnostic methods and solve bacteria Whether or not, the effect of reliable detection requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
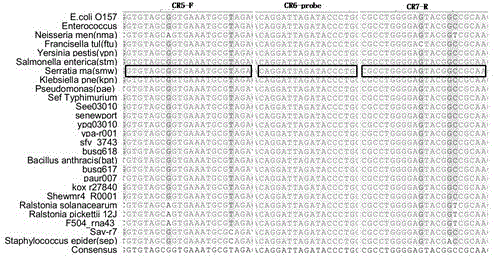
[0058] Example 1: Specificity of CR5-F and CR7-R primers for bacterial and human genomes
[0059] Dry powder primers and probes were dissolved in TE buffer to 10 μM; human genomic DNA and bacterial genomic DNA were extracted with kits, and quantified with a microspectrophotometer; conventional PCR was used for amplification, and the 30 μL reaction system included:
[0060] 2X Master Mix PCR Mix Buffer 15 μL
[0061] CR5-F upstream primer (10μM) 1 μL
[0062] CR7-R downstream primer (10μM) 1 μL
[0063]Genome template (50~100ng) 1 μL
[0064] Make up to 12 μL with double distilled water.
[0065] Reaction conditions: pre-denaturation at 95 °C for 5 min; denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, extension at 72 °C for 30 s, 30 cycles; final incubation at 72 °C for 5 min. PCR amplification products were detected by agarose gel electrophoresis, and the results were recorded, such as Figure 4 It was shown that this pair of primers amplified only the bacteri...
Embodiment 2
[0066] Example 2: Fluorescence quantitative PCR detects bacterial DNA
[0067] The embodiment does not use platelet product samples for detection, but uses human whole blood genome and bacterial genome alone or mixed to simulate, with human genome as a negative control, and mixed gradient dilution of human genome as a detection sample. Fluorescence quantitative PCR is performed. Detection, 30μL reaction system, including:
[0068] 2X Master Mix PCR Mix Buffer 15 μL
[0069] CR5-F upstream primer (10μM) 1 μL
[0070] CR7-R downstream primer (10μM) 1 μL
[0071] CR6-probe probe (10 μM) 0.5 μL
[0072] Internal reference probe (10μM) 0.5 μL
[0073] Genome template (50~100ng) 1 μL
[0074] Artificial reference (diluted to about 100 copies) 1 μL
[0075] Make up to 10 μL with double distilled water.
[0076] Reaction conditions: pre-denaturation at 95 °C for 2 min; denaturation at 95 °C for 20 s, annealing, extension and fluorescence detection at 60 °C for 1 min, a total of...
Embodiment 3
[0077] Embodiment 3: Difference and interpretation between the detection limit of artificial internal reference and the Ct value of sample detection
[0078] In this example, the artificial internal reference is synthesized in an amount of about 33ug per OD of the gene, and the number of molecules of the artificial internal reference is 38881.9, which is converted into a copy number for gradient dilution, and can be determined by fluorescence quantitative PCR under the conditions of this example (including reagents, Consumables, equipment, operation and other factors), the minimum number of internal parameters that can be detected is 20 copies. The OD of cultured Escherichia coli was detected by visible light spectrophotometry 600 value, in OD 600 When =1.0, there are about 6.3x10 in 1mL culture medium 8 50 μL of the dilution solution was used for lysing at 95°C for 20 minutes. After centrifugation, 1 μL of the diluted solution was used as a template for fluorescence quantit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com