A method for selectively extracting dna
A selective, DNA molecule technology, applied in the biological field, can solve the problems of low recovery efficiency, easy pollution, time-consuming and labor-intensive, etc., and achieve the effect of simple raw materials, cheap raw materials, and fast and convenient operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
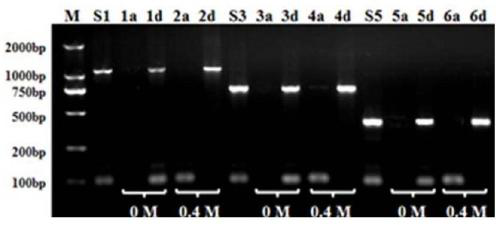
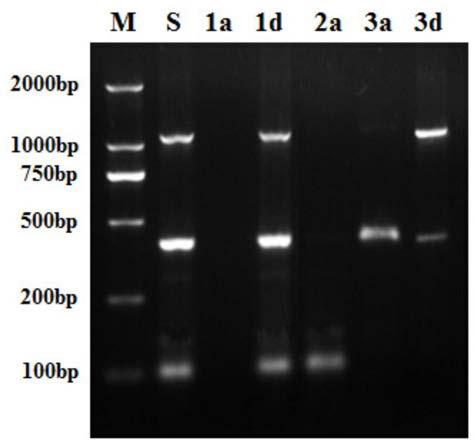
[0024] Disperse 0.5mg surface lysine-modified diatomaceous earth particles in 100μl aqueous solution containing 2000bp plasmid DNA and 100bp linear double-stranded DNA molecules (concentrations are both 25ng / μl), and use NaCl to adjust the system Na + The concentration is 0.4mol / L. Adjust the pH of the solution to 5.0, and shake the shaker for 20 minutes after mixing. Centrifugation at 6000 rpm was used to separate diatomaceous earth particles with DNA adsorbed. In addition, 0.75 mg of diatomaceous earth particles modified with surface lysine were dispersed in the adsorption supernatant, and the pH of the solution was adjusted to 4.0. After mixing, the shaker was shaken for 20 minutes, and the diatomaceous earth particles that had adsorbed DNA for the second time were separated by centrifugation at 6000 rpm.
[0025] The DNA-adsorbed diatomaceous earth particles separated by two centrifugation were separately dispersed in a pH=8.0 aqueous solution, and the shaker was shaken for ...
Embodiment 2
[0027] Disperse 1.0 mg of diatomaceous earth particles modified with surface lysine in 100 μl of an aqueous solution containing 2000 bp plasmid DNA and 100 bp linear double-stranded DNA molecules (with a concentration of 25 ng / μl), and use KCl to adjust the K in the system. + The concentration is 0.5mol / L. Adjust the pH of the solution to 4.0, and shake the table for 30 minutes after mixing. Centrifugation at 6000 rpm was used to separate diatomaceous earth particles with DNA adsorbed. In addition, 1.0 mg of diatomaceous earth particles modified with surface lysine were dispersed in the adsorption supernatant, and the pH of the solution was adjusted to 3.0. After mixing, the shaker was shaken for 20 minutes, and the diatomaceous earth particles that had adsorbed DNA for the second time were separated by centrifugation at 6000 rpm.
[0028] Disperse the DNA-adsorbed diatomaceous earth particles separated by centrifugation twice in an aqueous solution of pH=11.0, mix well and shak...
Embodiment 3
[0030] Disperse 0.5 mg of diatomaceous earth particles modified with surface lysine in 100μl of aqueous solution containing 1000bp and 100bp linear double-stranded DNA molecules (at a concentration of 25ng / μl each), adjust the salt concentration in the system to 0.4mol / L, adjust the solution The pH value is 5.0, and the shaker shakes for 20 minutes after mixing. Centrifuge at 6000 rpm to separate diatomaceous earth particles that have adsorbed DNA. In addition, 1.0 mg of diatomaceous earth particles modified with surface lysine were dispersed in the adsorption supernatant, and the pH value of the solution was adjusted to 4.0. After mixing, the shaker was shaken for 20 minutes, and the diatomaceous earth particles that had adsorbed DNA for the second time were separated by centrifugation at 6000 rpm.
[0031] Disperse the DNA-adsorbed diatomaceous earth particles separated by centrifugation twice in a pH=10.0 aqueous solution, mix well, and shake in a shaker for 20 minutes. Cent...
PUM
| Property | Measurement | Unit |
|---|---|---|
| size | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com