Method for dyeing DNA in cesium chloride density gradient mixed liquor through GelGreen dye
A technology of density gradient and mixed solution, which is applied in the field of molecular biology to achieve the effect of verification accuracy, improvement of detection sensitivity and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
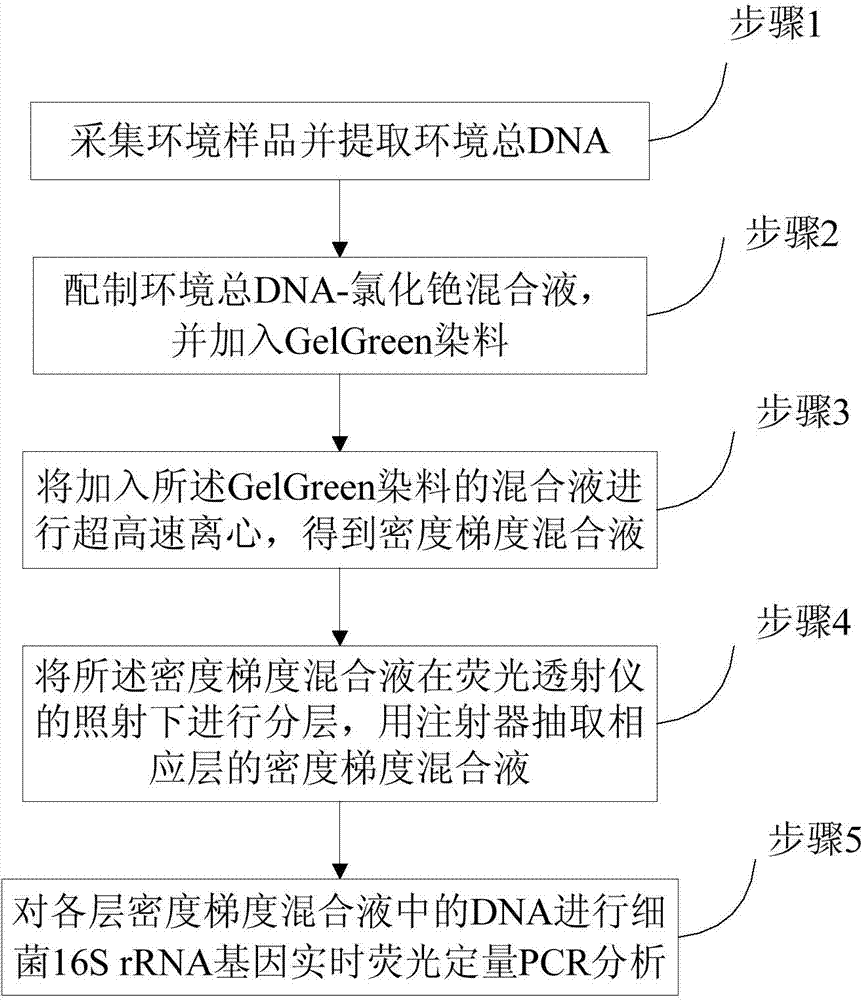
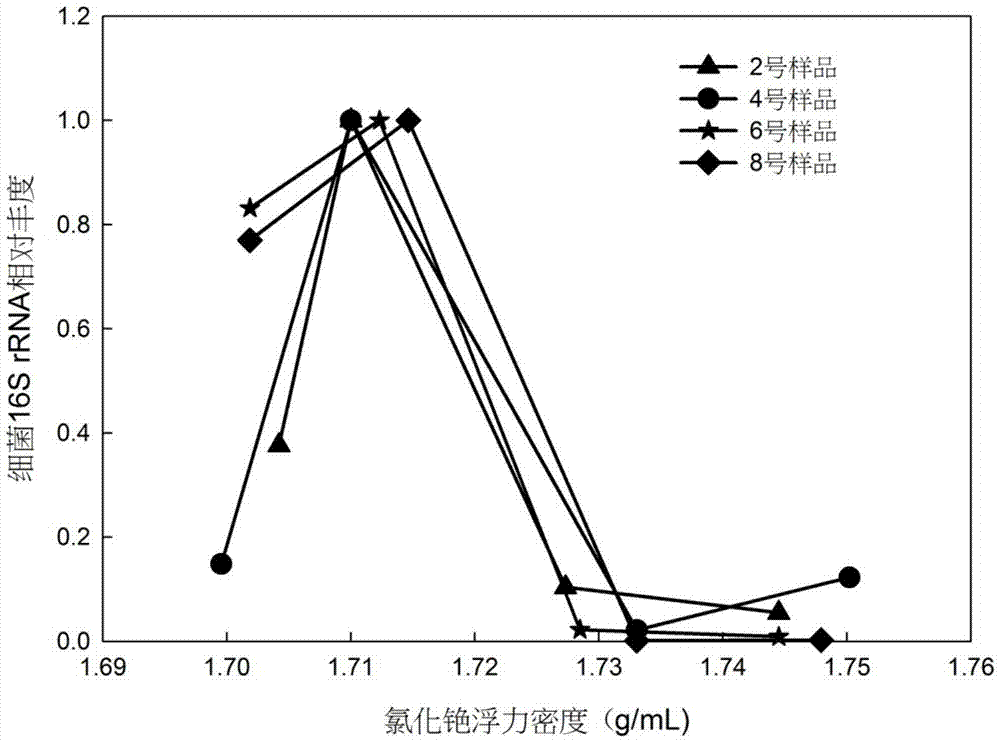
[0040] Prepare 1mL mixture of 5μg environmental total DNA and TE buffer, add 1.3g CsCl solid, vortex and mix well, add 4.7mL 1g / mL CsCl solution, add 8.85μL 10000X GelGreen dye to TE buffer treatment. The buoyant density of the prepared environmental total DNA-cesium chloride mixture was measured with an AR200 hand-held refractometer, and adjusted with 1g / mL CsCl solution and TE buffer, so that the buoyant density of the mixture was 1.72g / mL. Take 5.1 mL of the mixed solution and put it into an ultra-high-speed centrifuge tube, and centrifuge for 44 hours at 20°C and a centrifugal speed of 45,000 rpm. After the ultra-high-speed centrifugation, the density gradient mixture was irradiated by the fluorescence transilluminator, and the density gradient mixture in the ultra-high-speed centrifuge tube was divided into 4 parts based on the fluorescent DNA of the target layer. Layers I, II, III, and IV, in which the target band is in layer II, are drawn out sequentially from top to bo...
Embodiment 2
[0045] Prepare 5 μg of environmental DNA and GB buffer solution into 1 mL mixture, add 1.3 g of CsCl solid, vortex and mix well, then add 4.7 mL of 1 g / mL CsCl solution, and then add 8.85 μL of 10000X GelGreen dye to GB buffer solution. The buoyant density of the prepared environmental total DNA-cesium chloride mixture was measured with an AR200 hand-held refractometer, and adjusted with 1g / mL CsCl solution and GB buffer, so that the buoyant density of the mixture was 1.72g / mL. Take 5.1 mL of the mixed solution and put it into an ultra-high-speed centrifuge tube, and centrifuge for 44 hours at 20°C and a centrifugal speed of 45,000 rpm. After the ultra-high-speed centrifugation, the density gradient mixture was irradiated by the fluorescence transilluminator, and the density gradient mixture in the ultra-high-speed centrifuge tube was divided into 4 parts based on the fluorescent DNA of the target layer. Layers I, II, III, and IV, in which the target band is in layer II, are d...
Embodiment 3
[0051] Repeat the operation process of embodiment 1.
[0052] Observe the repeatability experiment results of DNA staining with GelGreen dye in TE buffer. In the repeatability experiment, the staining effect of GelGreen dye on DNA is consistent, indicating that the staining effect of GelGreen dye on DNA in the cesium chloride density gradient mixture has good repeatability.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com