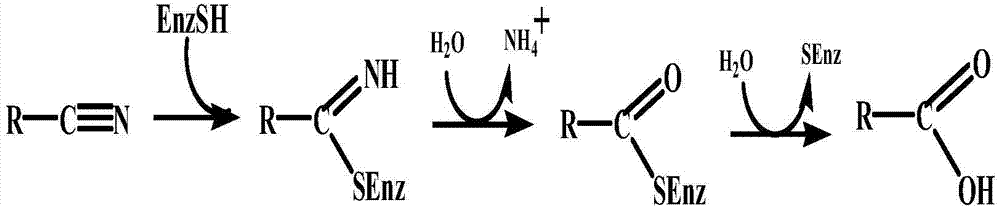
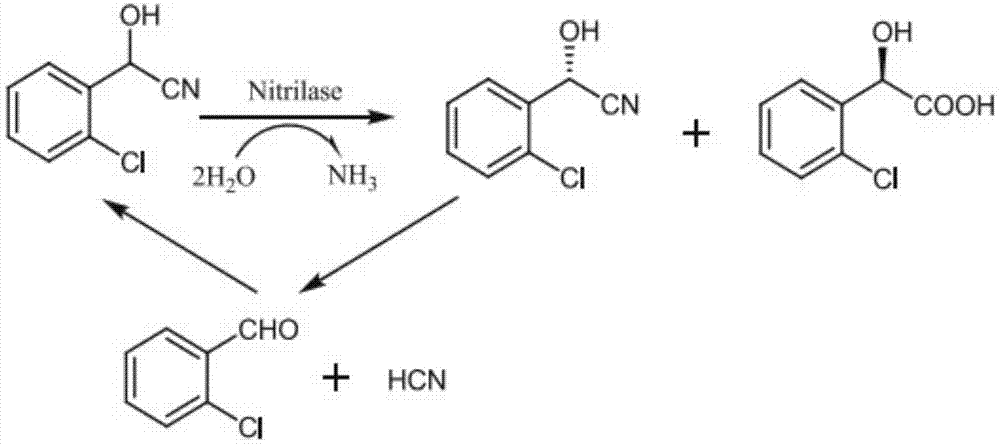
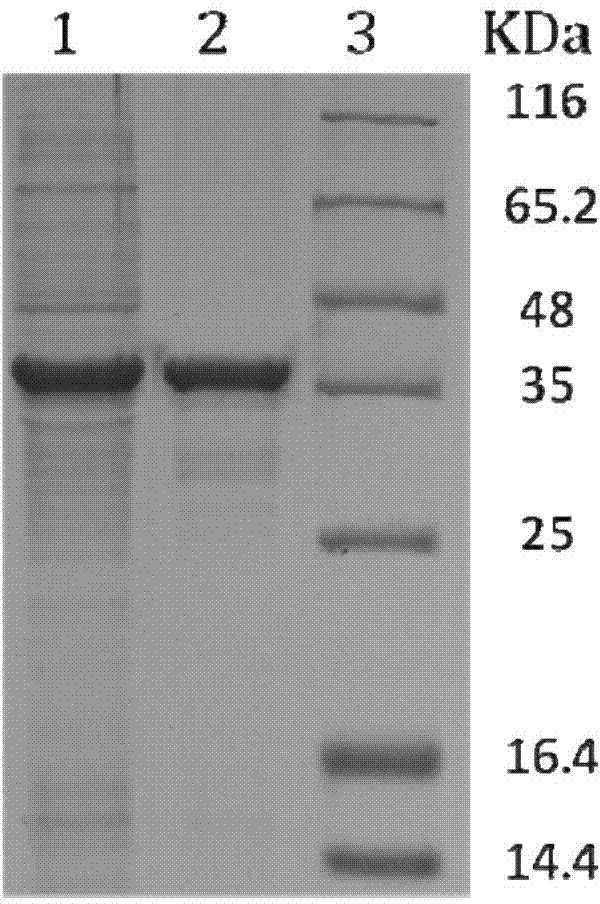
Nitrilase mutant and application thereof
A technology of nitrilase and mutants, applied in the direction of hydrolytic enzymes, enzymes, and the introduction of foreign genetic material using vectors, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Gene synthesis of parent nitrilase
[0033] In order to express the protein with the His-tag after the gene of the parental nitrilase (GeneBank: AY487562) was connected to the vector pET-28b, its stop codon was removed, and the codon preference of E. The sequence was sequence optimized and NcoI was introduced at the start codon to insert glycine after methionine. The sequence of the newly designed nitrilase gene (tegi) is shown in SEQ ID NO.1 (the amino acid sequence is shown in SEQ ID NO.2). The gene synthesis was entrusted to Shanghai Xuguan Biotechnology Development Co., Ltd. After the gene synthesis Linked to the expression vector pET-28b.
Embodiment 2
[0034] Construction of embodiment 2 mutant library
[0035] Using the parental sequence of the nucleotide sequence shown in SEQ ID NO.1 obtained in Example 1 as a template, the following two pairs of nucleotide sequences were used as primers to perform PCR amplification respectively, and the 132-threonine was mutated into the essence respectively. Amino acid single mutant Thr132-Arg (the nucleotide sequence is shown in SEQ ID NO.3, the amino acid sequence is shown in SEQ ID NO.4), the 189-position phenylalanine is mutated into the single mutant Phe189 of serine -Ser (the nucleotide sequence is shown in SEQ ID NO.5, the amino acid sequence is shown in SEQ ID NO.6) and the single mutant Phe189-Thr (nucleoside The acid sequence is shown in SEQ ID NO.7, and the amino acid sequence is shown in SEQ ID NO.8).
[0036] Two pairs of oligonucleotide primers are as follows (5'-3'):
[0037] Forward primer (132Thr): CTGAAACCGACCNNNGTGGAGCGTACCCTG
[0038] Reverse primer (132Thr): CAGGG...
Embodiment 3
[0048] Example 3: Screening of Mutant Library
[0049] (1) Primary screening of mutant library
[0050] Pick a single colony cultured in Example 2 and place it in a 96-well plate containing 1 mL of LB culture solution, add Kan (final concentration 50 μg / mL), and culture at 37°C until OD 600 When it reaches 0.6-0.8, add IPTG (final concentration 0.1mM) and induce at 28°C for 12h. Centrifuge at 9000rpm for 10min, remove the supernatant, and obtain bacterial cells. Pick wet bacteria and resuspend with 1mL (pH 7.5, 0.1M) phosphate buffer to prepare bacteria suspension. The reaction was carried out in an Ep tube, in a 1mL system: 900μl (pH 7.5, 0.1M) phosphate buffer (containing 5% methanol at the final volume concentration), 100μL of the above bacterial suspension, with a final concentration of 20mM o-chloromandelonitrile, at a temperature of 40°C , rotate at 1000 rpm, react for 5 min, and stop the reaction with 10 μl of 6M HCl. Enzyme activity in resting cells was measured us...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com