Molecular markers for haplotype identification of paddy rice CMS restoring gene Rf-1 segment and applications thereof
A molecular marker and gene restoration technology, applied in the field of plant biotechnology and molecular breeding, can solve the problems of chain drag and loss of target genes, and achieve the effect of overcoming the chain drag.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
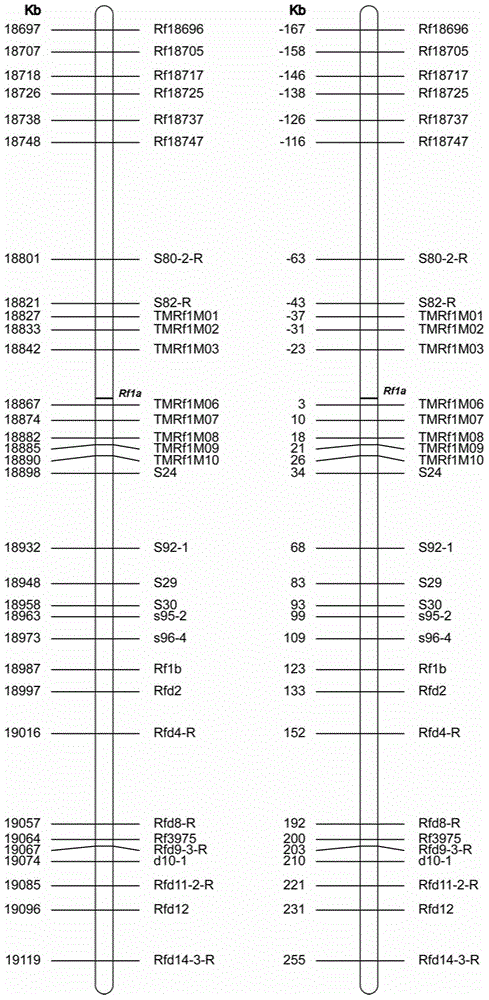
[0057] Example 1 Design of Molecular Marker of Rice CMS Restoration Gene Rf-1 Segment
[0058] Using next-generation sequencing technology (Illumina MiSeq sequencer), the parents of the rice wild-type and Baotai-type CMS restorer lines Minghui 63, the parent of the Honglian type CMS restorer line 9311, the self-developed excellent restorer line G527 and the rice wild-type restorer line The maintainer parent Zhenshan 97 was resequenced with an average 20-fold coverage of the whole genome, and the Rf-1 gene and its upstream and downstream regions of 200kb (genetic distance of about 1cM interval) were selected to discover the variation sites. The specific steps of marking design are as follows:
[0059] The first step is to compare the reference genome, compare the sequencing reads of each material to the Nipponbare genome, and use the software BWA for comparison. The reference genome sequence is the genome sequence of MSU version 6.1 Nipponbare.
[0060] The second step is to i...
Embodiment 2
[0063] Example 2 Preparation of Rice CMS Restoration Gene Rf-1 Segment OA Chip
[0064] Submit 128 sequences SEQ ID No.1-SEQ ID No.128 of 32 molecular markers obtained in Example 1 to Life Technologies to synthesize corresponding primers and TaqMan-MGB probes, and make an OpenArray chip, which can detect 96 simultaneously sample.
Embodiment 3
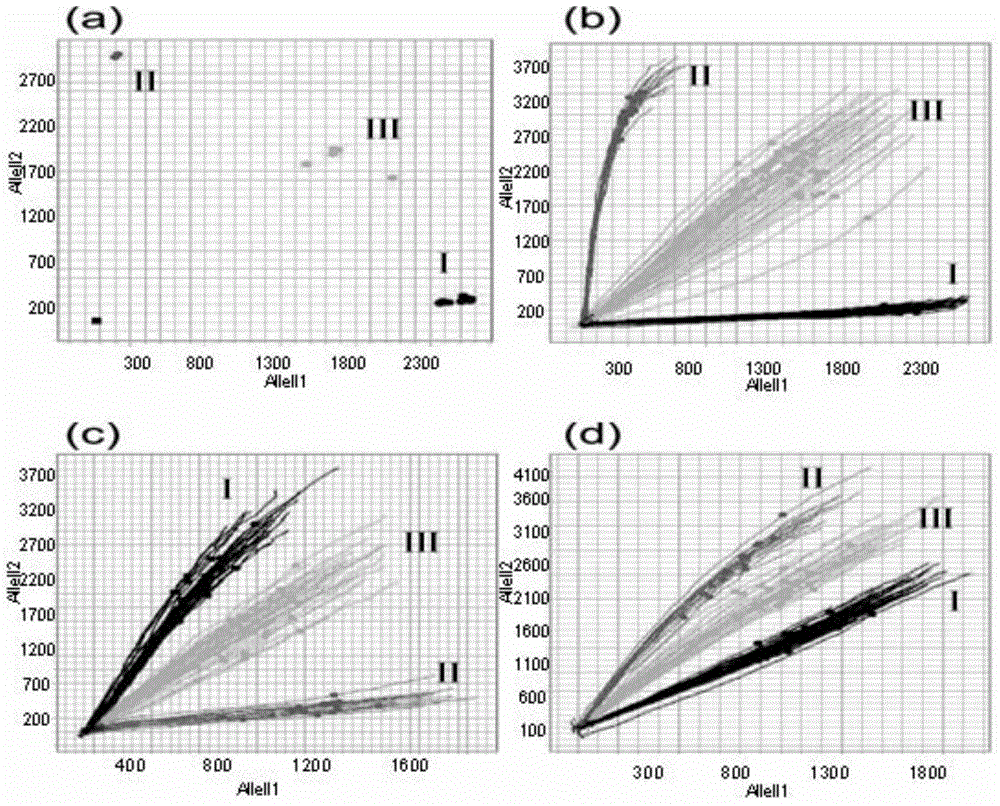
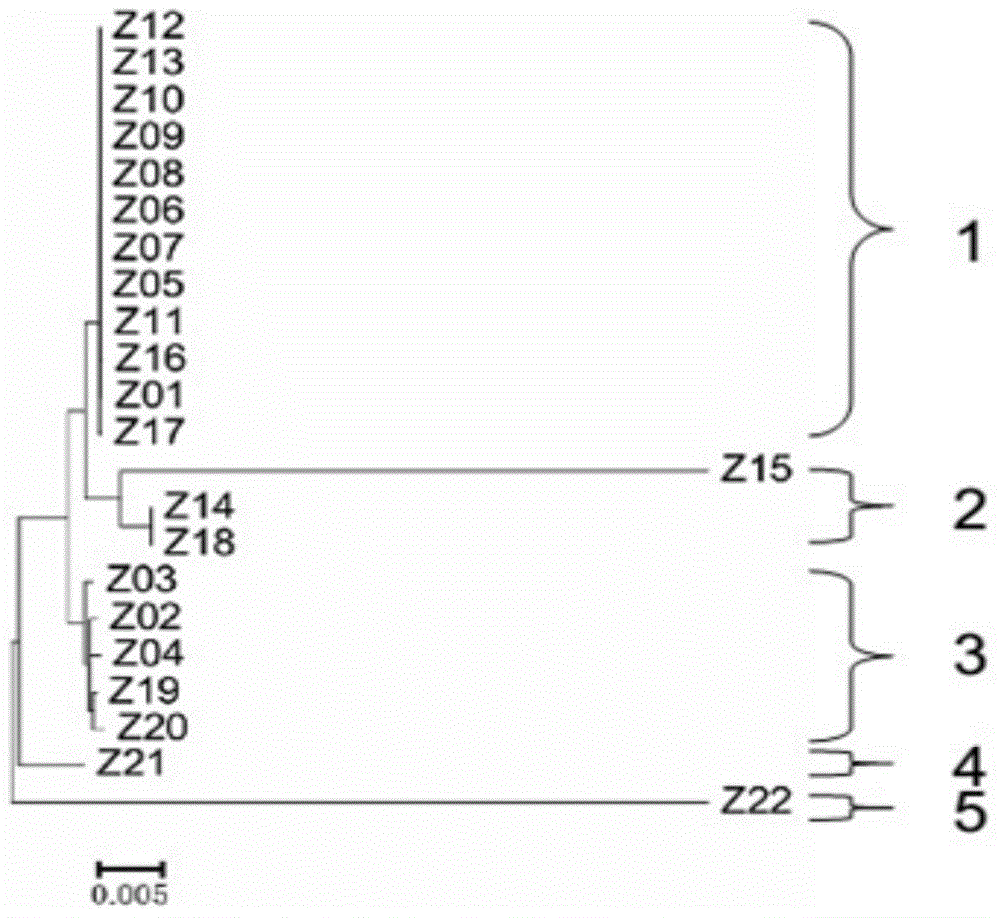
[0065] Example 3 Validation of the application effect of molecular markers for identifying the haplotype of the rice CMS restorer gene Rf-1 segment in the detection of rice DNA samples
[0066] In order to verify the haplotype identification method of the rice CMS recovery gene Rf-1 segment, it is necessary to test the primer probe combination, the actual detection and typing effect of the OA chip, and compare whether the design reference sample genotype is consistent with the actual detection genotype , so as to determine whether the primer probe set and OA chip have achieved the expected effect of design. The concrete steps of this embodiment are as follows:
[0067] 1. Rice genomic DNA extraction:
[0068] Genomic DNA was extracted from rice seeds, leaves and other tissues according to the detection requirements, and the DNA of young rice leaves was extracted using the standard procedure of the Promega Plant Genome Extraction Kit.
[0069] 2. DNA sample quality testing:
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com