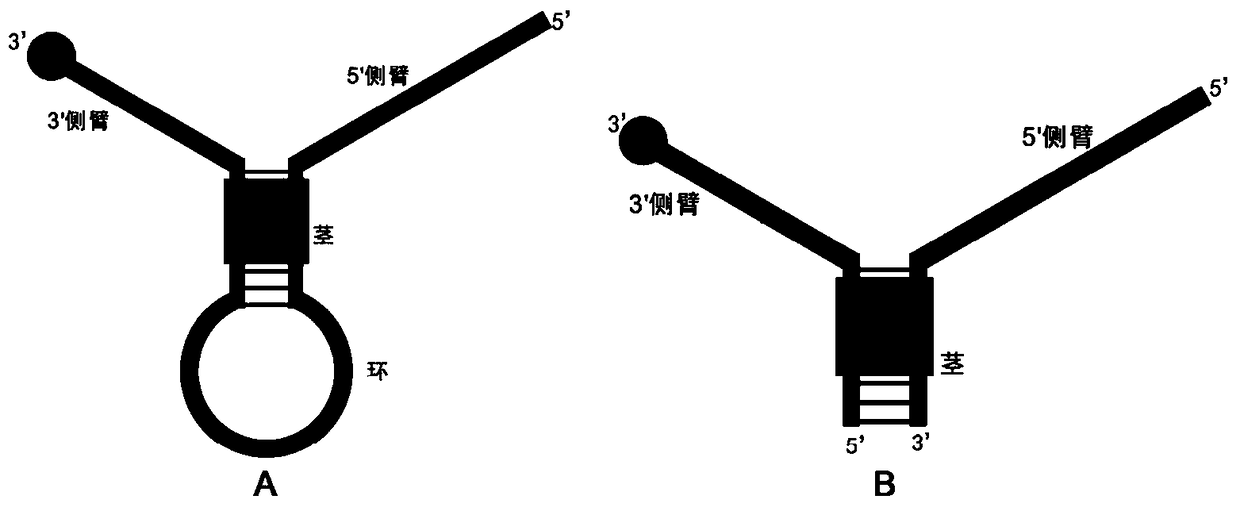
A nucleic acid isothermal amplification method based on positioning probe-mediated cleavage
A technology for isothermal amplification and positioning probes, applied in the field of thermal amplification, to achieve the effects of broadening the application range, strong versatility, high sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
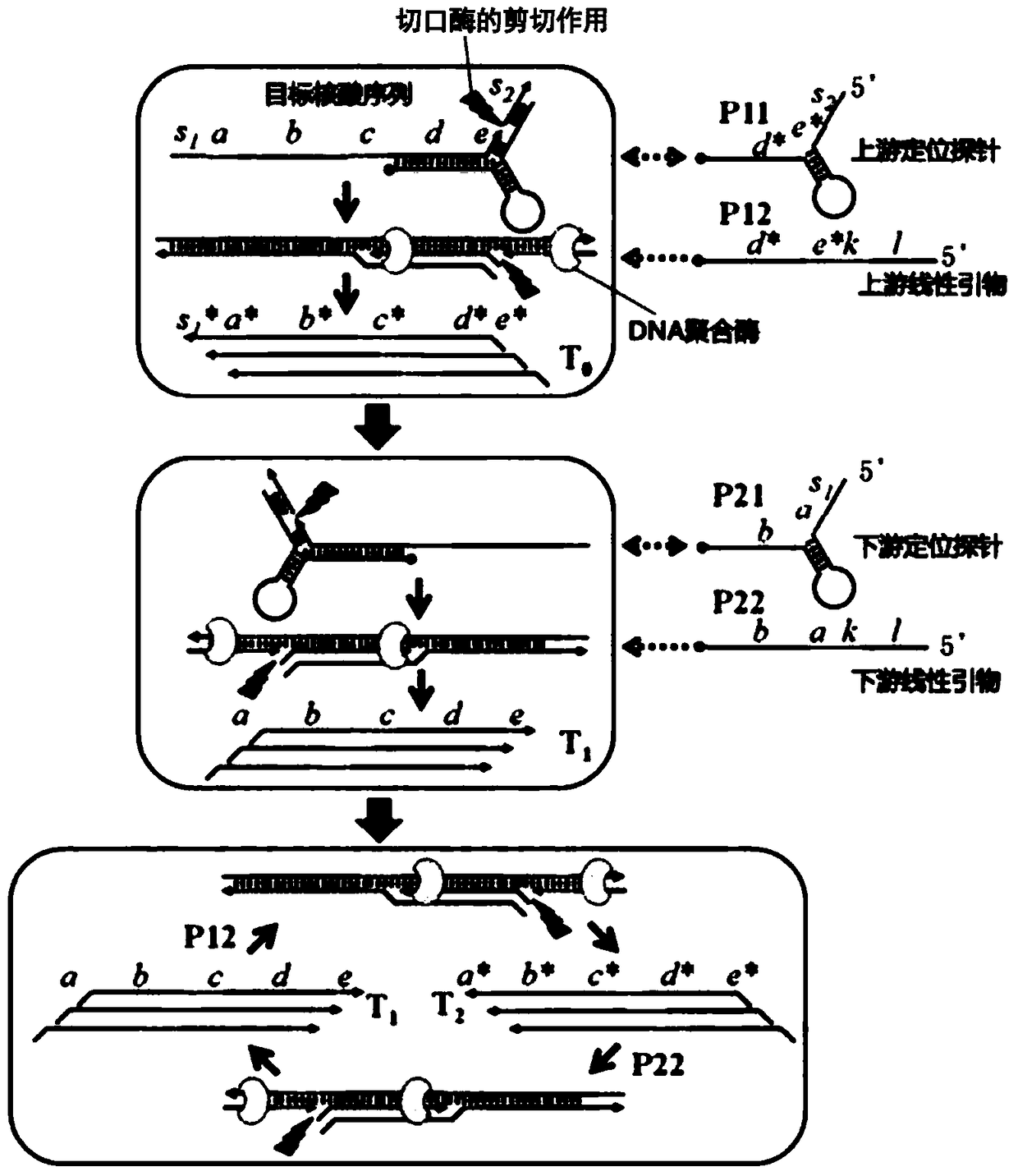
[0061] In this example, artificially synthesized target nucleic acid sequences were used to verify the feasibility of the method. According to the cleavage site and length requirements of the target nucleic acid sequence, corresponding positioning probes and primers are designed, and the amplified products are analyzed by agarose gel (3%) electrophoresis. Reaction principle see image 3 ,Specific steps are as follows:
[0062](1) Take 2 EP tubes, add 2.5μL reaction buffer (100mMNaCl, 50mM Tris-HCl, 60μg / mLBSA), 0.6μL endonuclease Nt.BstNBI (concentration: 10U / μL), 0.1μL Bst DNA polymerase (concentration is 8U / μL), 1μL dNTPs (concentration is 10mM), 4μL MgCl 2 (concentration is 25mM), 0.25 μL upstream localization probe P1 (concentration is 10 μM), 0.25 μL downstream localization probe P2 (concentration is 10 μM), 0.25 μL upstream primer L1 (concentration is 10 μM), 0.25 μL downstream primer L2 (concentration 10 μM) and 12.8 μL water, labeled a, b;
[0063] (2) Add 2.5 μL o...
Embodiment 2
[0078] This example is used to verify the amplification specificity of the method of the present invention. In this example, the analyte is an artificially synthesized target nucleic acid sequence (completely matched with the positioning probe / primer) and a mismatched nucleic acid sequence (only one base mismatch), according to the cleavage site and length of the target sequence It is required to design corresponding upstream and downstream positioning probes and upstream and downstream primers, and use SYBR Green I, a specific fluorescent dye dependent on double-stranded DNA, to display signal changes. For detection principle see image 3 ,Specific steps are as follows:
[0079] (1) Take 5 EP tubes, add 2.5μL reaction buffer (100mMNaCl, 50mM Tris-HCl, 60μg / mLBSA), 0.6μL endonuclease Nt.BstNBI (concentration: 10U / μL), 0.1μL Bst DNA polymerase (concentration is 8U / μL), 1μL dNTPs (concentration is 10mM), 4μL MgCl 2 (concentration is 25mM), 0.25 μL upstream localization probe ...
Embodiment 3
[0097] This example is used to verify the amplification sensitivity of the method of the present invention. In this example, artificially designed, synthetic target sequences were used to test the detection sensitivity of the method. According to the cleavage site and length requirements of the target sequence, the corresponding positioning probes and primers are designed, and the specific fluorescent dye SYBR Green I dependent on double-stranded DNA is used to display the signal change. For detection principle see image 3 ,Specific steps are as follows:
[0098] (1) Take 6 EP tubes, add 2.5μL reaction buffer (100mMNaCl, 50mM Tris-HCl, 60μg / mLBSA), 0.6μL endonuclease Nt.BstNBI (concentration: 10U / μL), 0.1μL Bst DNA polymerase (concentration is 8U / μL), 1μL dNTPs (concentration is 10mM), 4μL MgCl 2 (concentration is 25mM), 0.25 μL upstream localization probe P5 (concentration is 10 μM), 0.25 μL downstream localization probe P6 (concentration is 10 μM), 0.25 μL upstream prime...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com