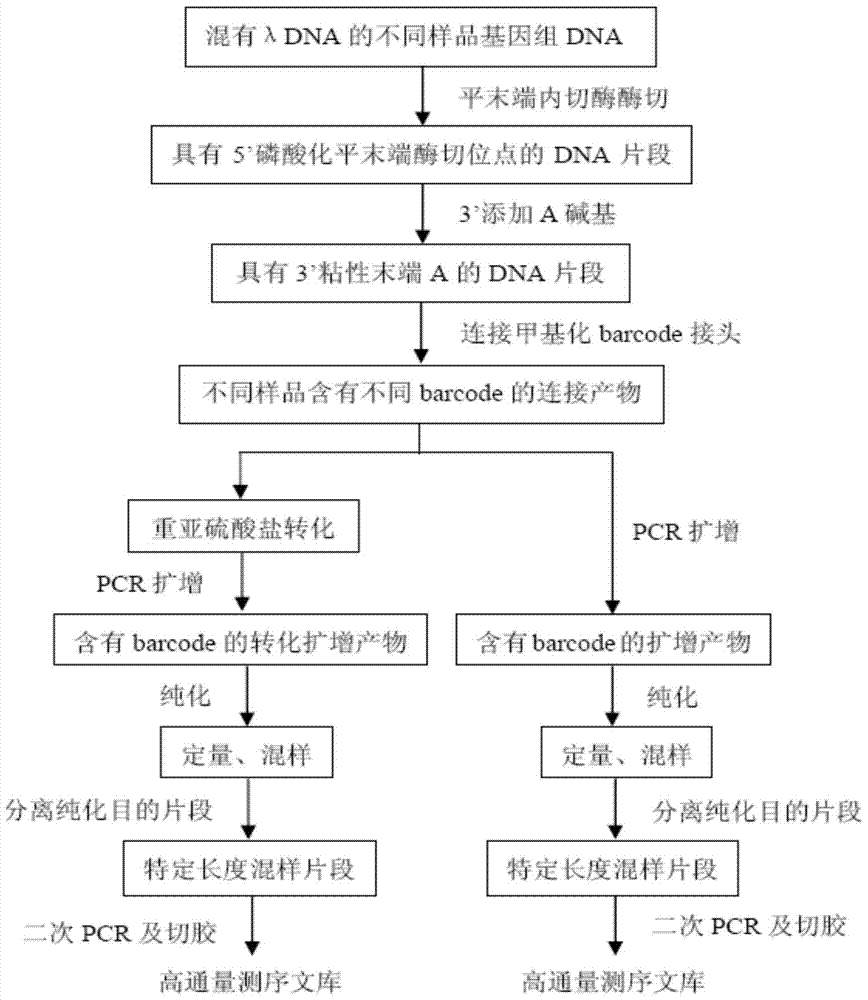
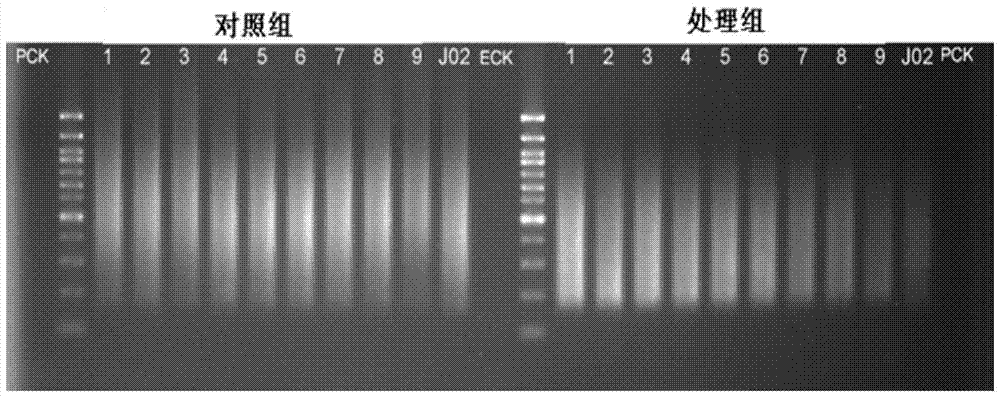
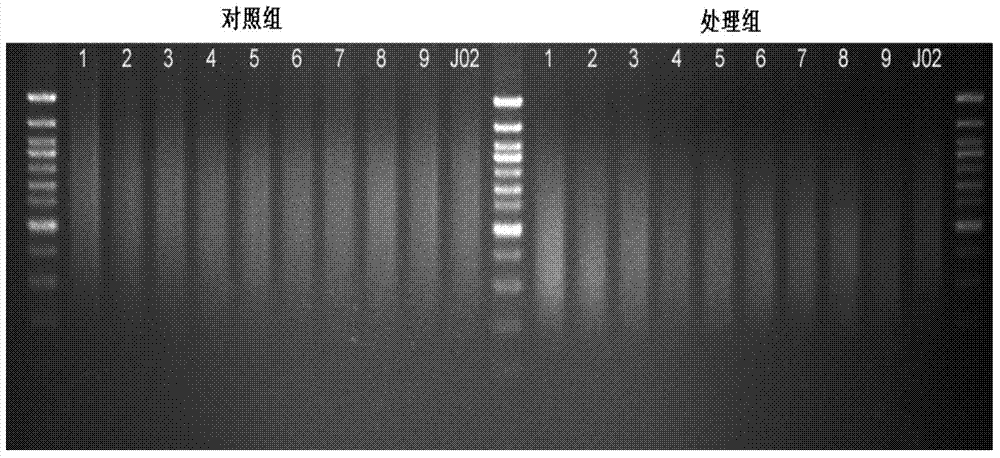
Construction method of high-throughput simplified methylation sequencing library without reference genome
A technology of reference genome and construction method, applied in the field of construction of simplified methylation sequencing library for species without reference genome, which can solve the problems of high cost of library construction and low efficiency of restriction endonuclease MspI digestion, and achieve simplified library preparation Steps to ensure consistent results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 Construction of a high-throughput simplified methylation sequencing library without a reference genome and its application to the epigenetic evolution analysis of Abies longbract
[0038]In this example, 9 individual samples of Abies georgei were selected as experimental materials, and the construction of a high-throughput simplified methylation sequencing library without a reference genome included the following steps:
[0039] 1. Genomic DNA of Abies longbract was diluted to 100ng / μL (Nanodrop quantification);
[0040] 2. Fragmentation of genomic DNA Use the blunt end restriction endonuclease HaeIII to digest the genomic DNA diluted in the first step. The enzyme digestion reaction system is:
[0041]
[0042]
[0043] The mixed system was incubated in a constant temperature water bath at 37°C for 5 hours.
[0044] 3. Add the reaction reagents required to produce the sticky A-terminus at the 3' end to the enzyme digestion reaction product in step 2, in...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com