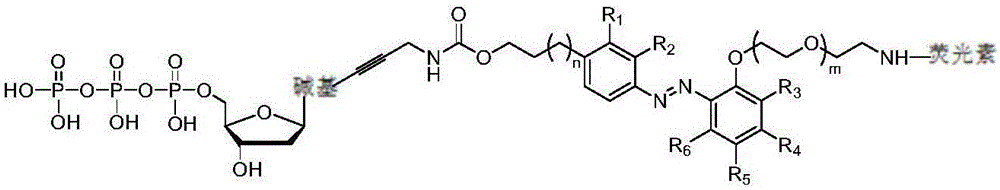
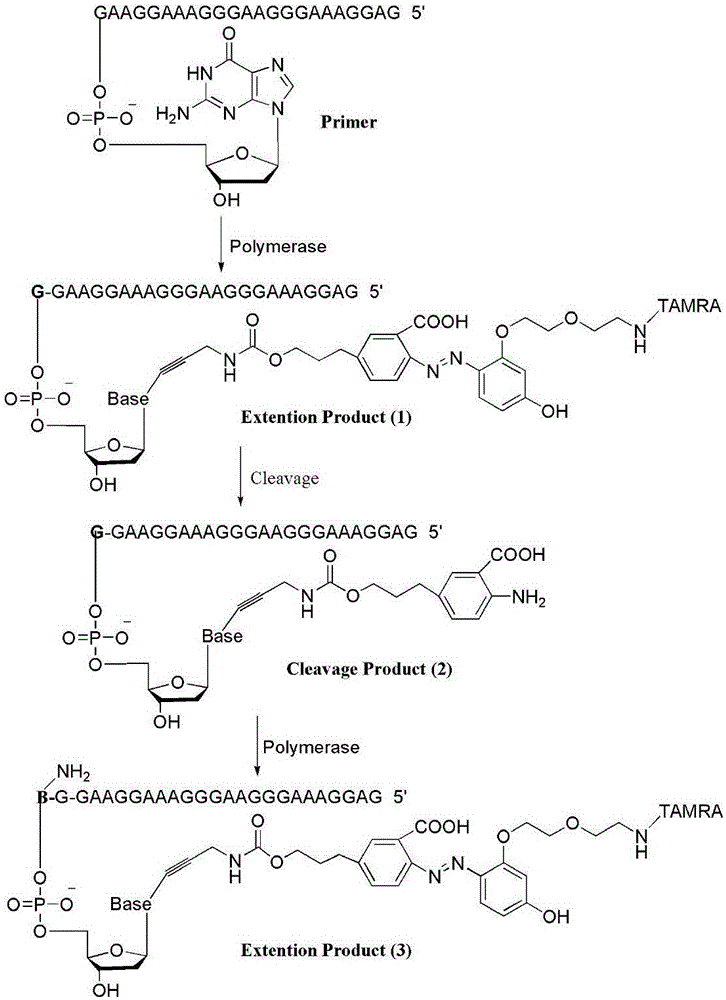
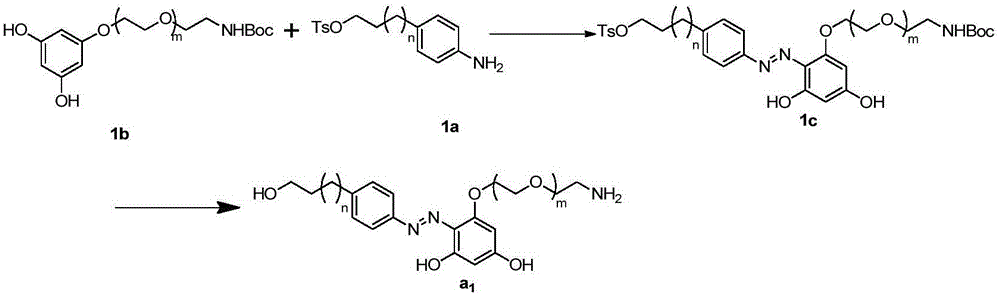
Fluorescence-labeled azo-modified nucleotide and application thereof in DNA sequencing
A fluorescent labeling and nucleotide technology, applied in the field of DNA sequencing, can solve the problems of insufficient shearing conditions, low efficiency, short read length, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Embodiment 1, azo linking unit (a 1 )Synthesis
[0075] The synthesis schematic diagram of the azo linkage unit in this embodiment is as follows image 3 As shown, where m is 2 and n is 1, the specific steps are as follows:
[0076] (1) Synthesis of 1c: 1a (710 mg, 2 mmol) was weighed and dissolved in 5 mL of water, and 0.5 mL of concentrated hydrochloric acid was slowly added in an ice bath. Sodium nitrite (145mg, 2.1mmol) was weighed and dissolved in 10mL of water, slowly added dropwise to the above reaction solution at 0°C, and stirred for 40min. Weigh 1b (694mg, 2mmol) and sodium hydroxide (80mg, 2mmol), add 15mL ethanol and 45mL water into an ice-water bath and stir to dissolve. The generated diazonium salt was slowly added dropwise to the aqueous sodium hydroxide solution of 1b in an ice-water bath, stirred for 5 hours, and a large amount of precipitate was formed. After filtration, the solid was washed three times with 10 mL of water, and dried to obtai...
Embodiment 2
[0078] Embodiment 2, azo linking unit (a 2 )Synthesis
[0079] The synthesis schematic diagram of the azo linkage unit in this embodiment is as follows Figure 4 As shown, where m is 2 and n is 1, the specific steps are as follows:
[0080] (1) Synthesis of 2c: 1a (710 mg, 2 mmol) was weighed and dissolved in 5 mL of water, and 0.5 mL of concentrated hydrochloric acid was slowly added in an ice bath. Sodium nitrite (145mg, 2.1mmol) was weighed and dissolved in 10mL of water, slowly added dropwise to the above reaction solution at 0°C, and stirred for 40min. Weigh 2b (682 mg, 2 mmol) and sodium hydroxide (80 mg, 2 mmol), add 15 mL of ethanol and 45 mL of water into an ice-water bath and stir to dissolve. The generated diazonium salt was slowly added dropwise to the aqueous sodium hydroxide solution of 2b in an ice-water bath, stirred for 4 hours, and a large amount of precipitate was formed. After filtration, the solid was washed three times with 20 mL of water, and d...
Embodiment 3
[0082] Embodiment 3, azo linking unit (a 3 ) and (a 4 )Synthesis
[0083] The synthesis schematic diagram of the azo linkage unit in this embodiment is as follows Figure 5 As shown, where m is 2 and n is 1, the specific steps are as follows:
[0084] (1) Synthesis of 3c: 2a (814 mg, 2 mmol) was weighed and dissolved in 10 mL of water, and 0.5 mL of concentrated hydrochloric acid was slowly added in an ice bath. Sodium nitrite (145mg, 2.1mmol) was weighed and dissolved in 10mL of water, slowly added dropwise to the above reaction solution at 0°C, and stirred for 40min. Weigh 2b (682 mg, 2 mmol) and sodium hydroxide (80 mg, 2 mmol), add 15 mL of ethanol and 45 mL of water into an ice-water bath and stir to dissolve. The generated diazonium salt was slowly added dropwise to the aqueous sodium hydroxide solution of 2b in an ice-water bath, stirred for 4 hours, and a large amount of precipitate was formed. After filtration, the solid was washed three times with 20 m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com