Application of DNA three-way regulation activation based hybridization chain reaction to high sensitivity detection of DNA methyltransferase
A hybrid chain reaction and methylation technology, applied in the field of enzyme activity detection, can solve problems such as lack of recognition mechanism, and achieve good specificity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Embodiment one, experimental method
[0038] The role of M.SssI and its recognition sequence
[0039] Take different concentrations of M.SssI, 160μMSAM and methyltransferase recognition probe and mix with NEBuffer2 (1×), react for 2 hours, add methylation-sensitive restriction endonuclease HpaII and mix with Cutsmart (1×) buffer , Reaction 2h. For HpaII inactivation, the inactivation condition is to keep at 80° C. for 30 minutes, and lower the temperature slowly.
[0040] Performing a hybridization chain reaction
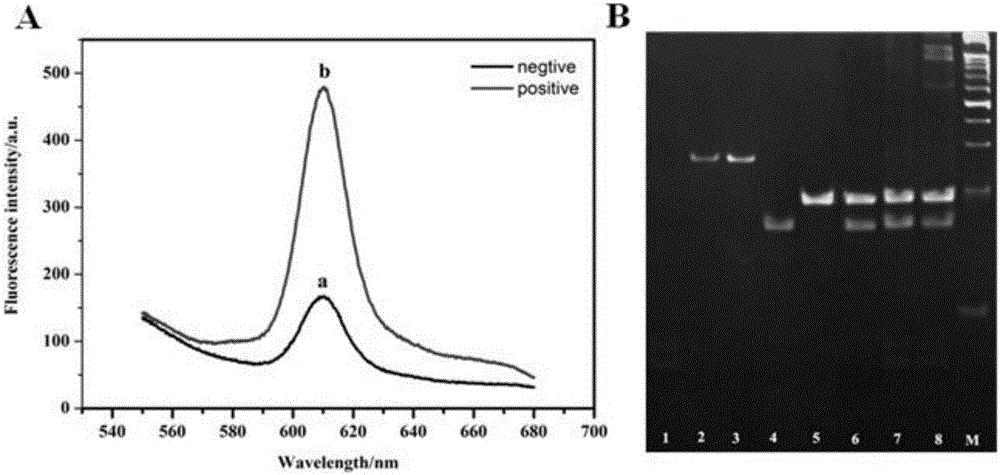
[0041] In the above system, add 1×TNaK buffer solution, H1, H2 and ultrapure water, vortex and mix well and react for 2 hours. Maintain the complete methyltransferase recognition sequence and mix with H1 and H2 for hybridization chain reaction.
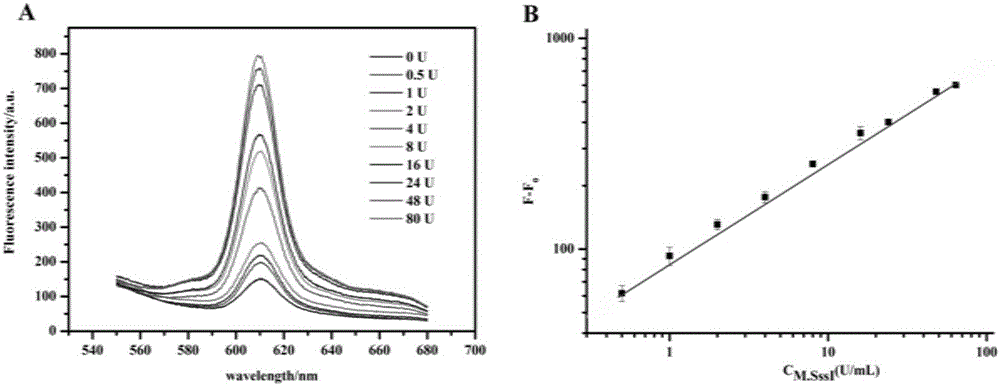
[0042] Dye addition and fluorescence measurement
[0043] After the above hybridization chain reaction is completed, add NMM, KCl and ultrapure water to the reaction system to increase its volume to 50 μL. Afte...
Embodiment 2
[0047] Example 2. The hybridization chain reaction based on DNA three-way joint activation is used for the highly sensitive detection of DNA methyltransferase
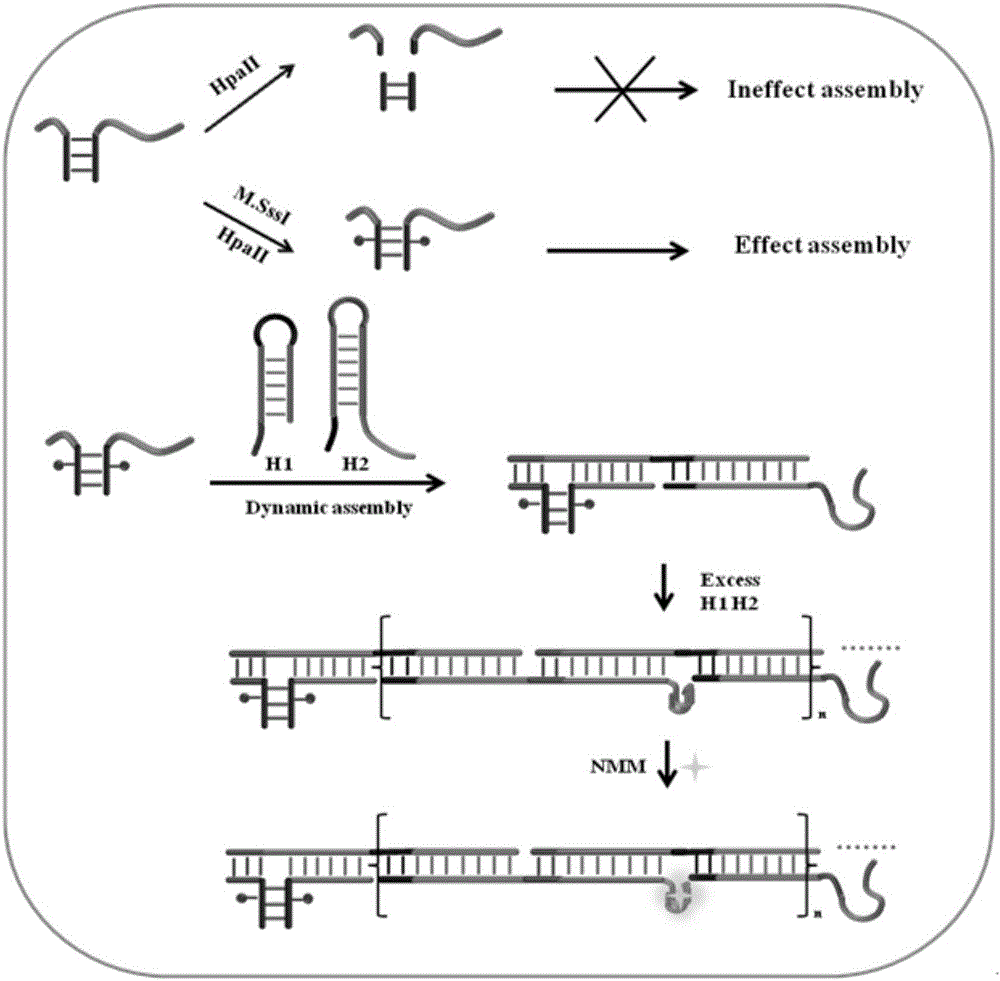
[0048] The Principle of Hybridization Chain Reaction Based on Activation of DNA Three-way Knot for Detection of M.SssI Activity
[0049] In order to achieve the generality of this method, we designed a novel DNA three-way junction activation method based on the protection mechanism of restriction endonucleases for the determination of CpG methyltransferase activity. Specific principles such as figure 1 Shown: The recognition probe (RMP) of M.SssI is formed by direct annealing of a DNA strand carrying a Toehold part and a DNA strand carrying a strand migration part. In RMP, the Toehold part and the strand migration part are close to each other, so they can act as the priming strand to trigger DNA assembly. In addition, its stem contains recognition sequences and sites for restriction endonucleases HpaII and M.SssI. W...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com