cDNA of chalcone isomerase that controls anthocyanin synthesis in Cymbidium japonica
A technology of chalcone isomerase and snow orchid, applied in the field of cDNA, can solve the problem of lack of freshness of monocotyledons
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 wxya Cloning of full-length genes
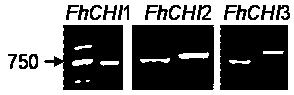
[0034] wxya Acquisition of full-length gene sequence: Design specific upstream and downstream primers based on known transcriptome sequencing results. Use the Trizol kit to extract the total RNA of the petals of Cymbidium safflower, and reverse transcribe to generate the first strand of cDNA, which is used as a template for RT-PCR to obtain wxya The full-length sequence of the gene, the results are as follows Figure 1 .
[0035] The specific primers and specific PCR reaction conditions for cloning the full-length cDNA are as follows:
[0036] Primers CHIF1: TCTAATGACAACAACAACGC CHIR1: TTCTTACGACGTGATCCTTT
[0037] PCR conditions: pre-denaturation at 94°C for 10min; denaturation at 94°C for 30s; annealing at 56°C for 30s;
[0038] Extension at 72°C for 1min; final extension at 72°C for 10min; denaturation to extension for a total of 30 cycles.
[0039] Primer CHIF2: TTCTGATGACAATGGTGGAG CHIR2: TTTCTCCTCTGCCTTCCATA ...
Embodiment 2
[0045] Example 2 Homology Comparison Analysis
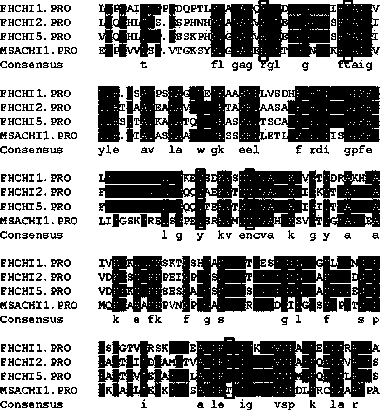
[0046] Express the protein sequence of FhCHIs and the protein sequence of alfalfa MsaCHI1 in FASTA format, save it as a TXT file, and then input it into ClustalW2 online software for homology comparison of their amino acids, and you can observe their most homologous amino acid positions; Put the protein sequence into the DNAMAN software according to the comparison results, and the identical amino acids are displayed with a dark blue background. Observe the four active sites of MsaCHI1, arginine (Arg36), threonine (Thr48), tyrosine (Tyr106 ), asparagine (Asn113), whether FhCHI1, FhCHI2, and FhCHI5 also have these amino acids.
Embodiment 3
[0047] Example 3 wxya Semi-quantitative expression analysis in different periods and its correlation with anthocyanin content
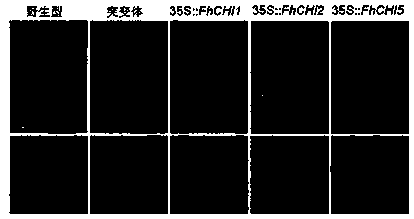
[0048] The RNAs of five different stages of safflower, pandan snow flower, green bud, red and green, red bud, first bloom, and full bloom were extracted respectively, and the total cDNA was obtained after inversion. Then, using it as a template, the primers of the three genes were used for semi-analysis. Quantitative RT-PCR was used to analyze the expression level. Using SPSS software, the gene expression level and the anthocyanin accumulation content in these five periods were correlated with each other.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com