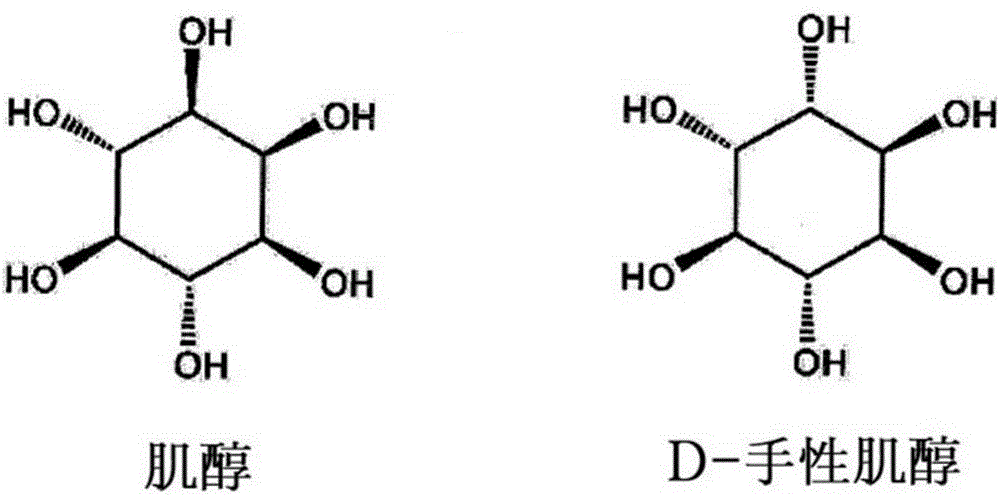
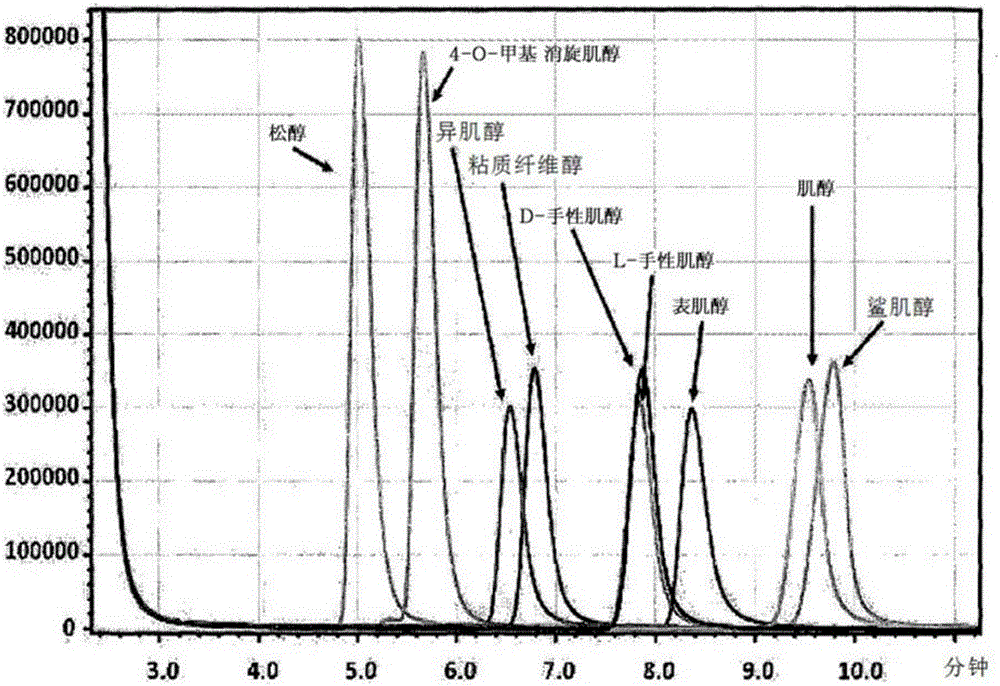
Method for preparing D-chiro-inositol using microbes
A chiral inositol and inositol technology, applied in biochemical equipment and methods, chemical instruments and methods, and the use of vectors to introduce foreign genetic material, etc., can solve problems such as high price, poor economy, and difficult to separate by-products
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Example 1: Cloning of the inositol transporter gene and preparation of a recombinant vector comprising the transporter
[0073] It has been reported that Bacillus subtilis, Salmonella typhimurium, and Agrobacterium tumefaciens, which use inositol as a carbon source, respectively cloned the major of myo-inositol transporter. Genes and minor genes.
[0074] In Bacillus subtilis (B.subtilis), the inositol transporter genes are iolT gene and iolF gene; in Salmonella typhimurium (S.typhimurium), the inositol transporter genes are iolT1 gene and iolT2 gene; In Bacillus (A. tumefaciens), the inositol transporter genes are Atu5935 gene and Atu2525 gene. The information of the above genes is shown in Table 1.
[0075] Table 1
[0076]
[0077] 1-1. Preparation of recombinant vectors pACYCD-BsiolT(F2) and pACYCD-BsiolT-BsiolF(F2)
[0078] In the case of the B. subtilis strain, the major transporter iolT and the minor transporter iolF were introduced into pACYCDuet-1 from t...
Embodiment 2
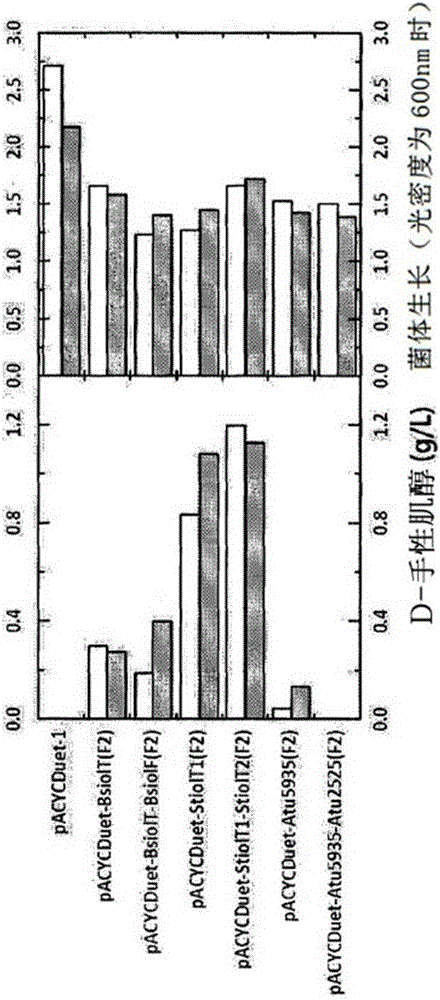
[0091] Example 2: Evaluation of the activity of the inositol transporter
[0092] As prepared in the above Example 1, each recombinant plasmid containing the inositol transporter derived from Bacillus subtilis (B. subtilis), Salmonella typhimurium (S. typhimurium), Agrobacterium tumefaciens (A. tumefaciens) , pACYCD-BsiolT(F2), pACYCD-BsiolT-BsiolF(F2), pACYCD-StiolT1(F2), pACYCD-StiolT1-StiolT2(F2), pACYCD-Atu5935(F2), pACYCD-Atu5935-Atu2525(F2) and containing The recombinant plasmid pCOLAD-sAtiep-sAtiepf, which converts inositol into D-chiro-inositol and the gene encoding inositol dehydrogenase and inositol isomerase, is transformed into Escherichia coli (E.coli) BL21 (DE3) together to prepare transformed strains.
[0093] 5 mL of the prepared recombinant transformed strain was cultured in a glass test tube with a diameter of 25 mm and a height of 150 mm. The culture conditions are as follows: utilize the M9 minimum culture medium that contains the inositol of 1% (w / v), th...
Embodiment 3
[0095] Example 3: Cloning of inositol dehydrogenase and inositol isomerase genes and preparation of recombinant vectors comprising them
[0096] Inositol is converted to D-chiro-inositol through the continuous reaction of the following reaction formula 1 to reaction formula 3. Inositol dehydrogenase (inositoldehydrogenase) catalyzes the reactions of reaction formula 1 and reaction formula 3, and inososeisomerase (inososeisomerase) catalyzes the reaction of reaction formula 2.
[0097] [Reaction 1]
[0098] Inositol + nicotinamide adenine dinucleotide (oxidized state) (NAD + ) 2-keto-inositol + nicotinamide adenine dinucleotide (reduced state) (NADH) + H +
[0099] [Reaction 2]
[0100] 2-Keto-Inositol 1-Keto-D Chiral Inositol
[0101] [reaction formula 3]
[0102] 1-Keto-D-chiro-inositol + nicotinamide adenine dinucleotide (reduced state) (NADH) + H + D-chiro-inositol + nicotinamide adenine dinucleotide (oxidized state) (NAD + )
[0103] Cloned from A. tumefaciens (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com