Method for expression of CRISPR sgRNA by eukaryotic cell III-type promoter and use thereof
A technology of promoter and RNA interference, applied in DNA/RNA fragments, recombinant DNA technology, introduction of foreign genetic material using vectors, etc., can solve problems such as unreported, and achieve the effect of simple structure and convenient construction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1 Target site selection
[0024] Based on the characteristics of the target sites of the CRISPR / Cas9 system, the target sites containing PAM (NGG / NGGNG) were searched in the genome. After screening on the CRISPRDesign website (http: / / crispr.mit.edu / ), the following two sites were selected as target sites:
[0025] VEGF Target Site Fragment:
[0026] CTCGGCCACCACAGGGAAGCTGG (SEQ ID NO: 1)
[0027] CCR5a / CCR2 target site fragment:
[0028]CACACTTGTCACCACCCCAAAGGTG (SEQ ID NO: 2)
Embodiment 2Ca
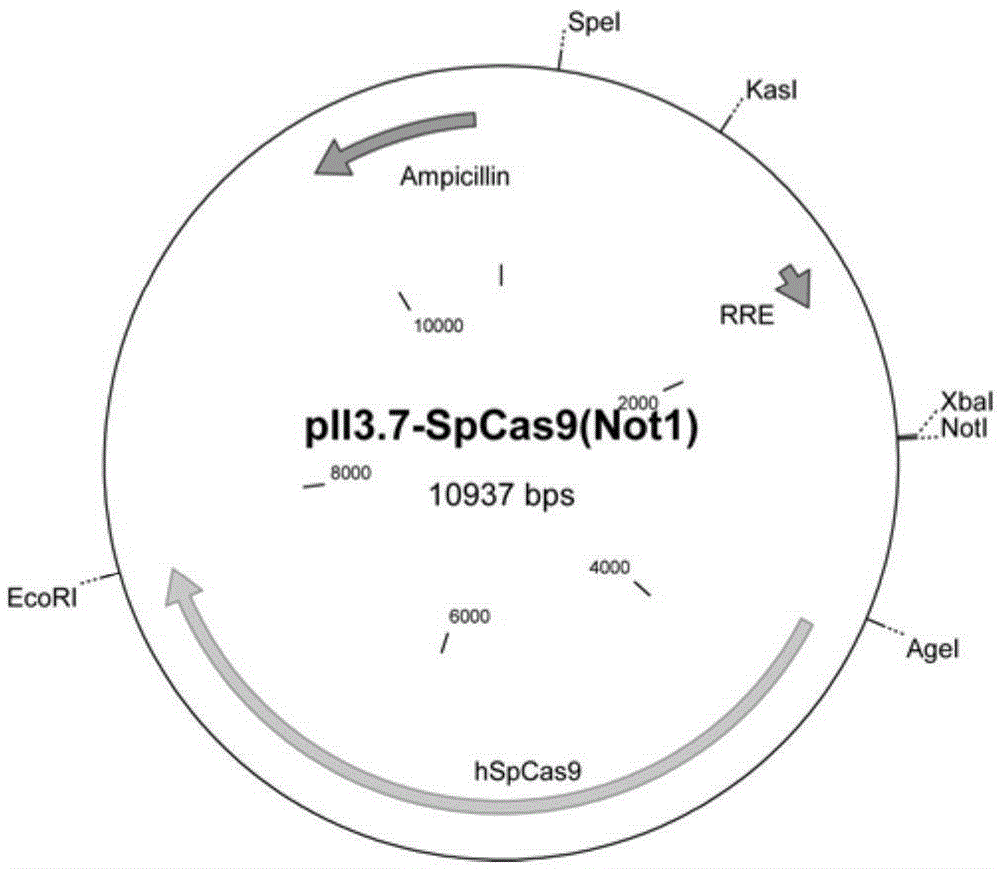
[0029] Example 2 Cas9 protein expression vector
[0030] The optimized Streptococcus pyogenes Cas9 sequence (hSpCas9) was inserted into the backbone vector pll3.7. like figure 2 shown. The sequence of hSpCas9 is as follows (SEQ ID NO: 3):
[0031] GACAAGAAGTACAGCATCGGCCTGGACATCGGCACCAACTCTGTGGGCTGGGCCGTGATC
[0032] ACCGACGAGTACAAGGTGCCCAGCAAGAAATTCAAGGTGCTGGGCAACACCGACCGGCAC
[0033] AGCATCAAGAAGAACCTGATCGGAGCCCTGCTGTTCGACAGCGGCGAAACAGCCGAGGCC
[0034] ACCCGGCTGAAGAGAACCGCCAGAAGAAGATACACCAGACGGAAGAACCGGATCTGCTAT
[0035] CTGCAAGAGATCTTCAGCAACGAGATGGCCAAGGTGGACGACAGCTTCTTCCACAGACTG
[0036] GAAGAGTCCTTCCTGGTGGAAGAGGATAAGAAGCACGAGCGGCACCCCATCTTCGGCAAC
[0037] ATCGTGGACGAGGTGGCCTACCACGAGAAGTACCCCACCATCTACCACCTGAGAAAGAAA
[0038] CTGGTGGACAGCACCGACAAGGCCGACCTGCGGCTGATCTATCTGGCCCTGGCCCACATG
[0039] ATCAAGTTCCGGGGCCACTTCCTGATCGAGGGCGACCTGAACCCCCGACAACAGCGACGTG
[0040] GACAAGCTGTTCATCCAGCTGGTGCAGACCTACAACCAGCTGTTCGAGGAAAACCCCATC
[0041] AACGCCAGCGGCGTGGACGCCAAGGCCATC...
Embodiment 3
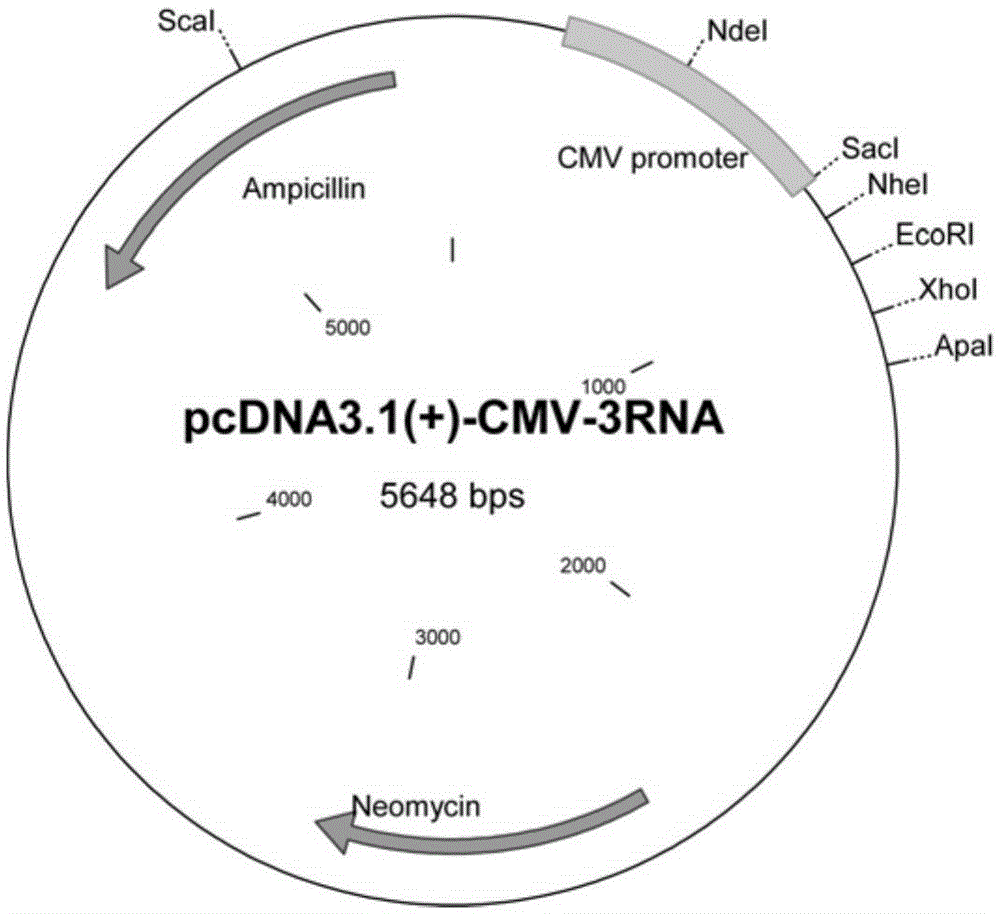
[0100] Example 3 Multiple small RNA expression vectors (msgRNA-2) connected in series
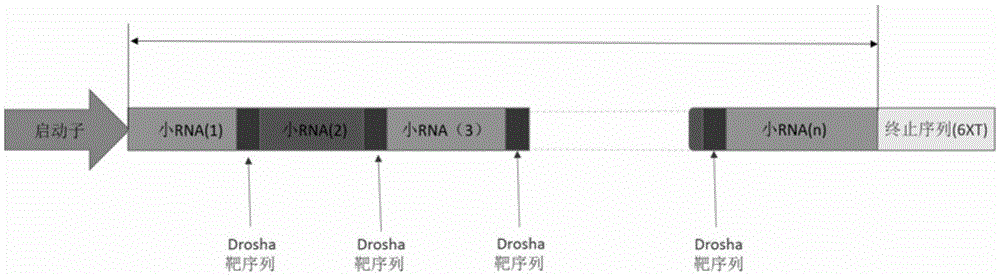
[0101] Taking the experimental carrier as an example, the carrier is designed as a U6-sgRNA-shRNA-sgRNA structure. According to the selected two target sites (VEGF target site and CCR5a target site), the corresponding sgRNA sequences are respectively designed, with CD40 shRNA sequences of Drosha target sites. The individual fragments are joined together by cutting sites of different restriction endonucleases. The sequences of the two Drosha target sites used in this experiment are as follows:
[0102] Drosha target site fragment 1: TCCGAGGCAGTAGGCA (SEQ ID NO: 4)
[0103] Drosha target site fragment 2: TGCTGTTGACAGTGAGCG (SEQ ID NO: 5)
[0104] Construction of this vector requires two steps. In the first step, the gene fragments of the three RNA sequences were inserted between the multiple cloning sites NheI and ApaI on the backbone vector pcDNA3.1(+) (Invitrogen), and each fragment was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com