HRM method for detecting genetic polymorphism of CYP3A4*1G and MDR1C1236T
A technology of MDR1C1236T and gene polymorphism, applied in the field of biotechnology and medicine, can solve the problems of long detection cycle, unsuitable for large sample detection, heavy workload, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0106] Example 1 Screening of CYP3A4*1G primers
[0107] Taking the CYP3A4*1G gene as an example, PrimerPremier software was used for primer design.
[0108] Sequence of CYP3A4*1G gene:
[0109] (SEQ ID NO11)
[0110] GGCTATGAAACCACGAGCAGTGTTTCTCCTTCATTATGTATGAACTGGCCACTCACCCTGATGTCCAGCAGAAACTGCAGGAGGAAATTGATGCAGTTTTACCCAATAAGGTGAGTGGATG R TACATGGAGAAGGAGGGAGGAGGTGAAACCTTAGCAAAAATGCCTCCTCACCACTTCCCAGGAGAA
[0111] ( R Indicates the gene mutation site)
[0112] The size of the primer fragment will affect the specificity of PCR amplification as follows:
[0113]
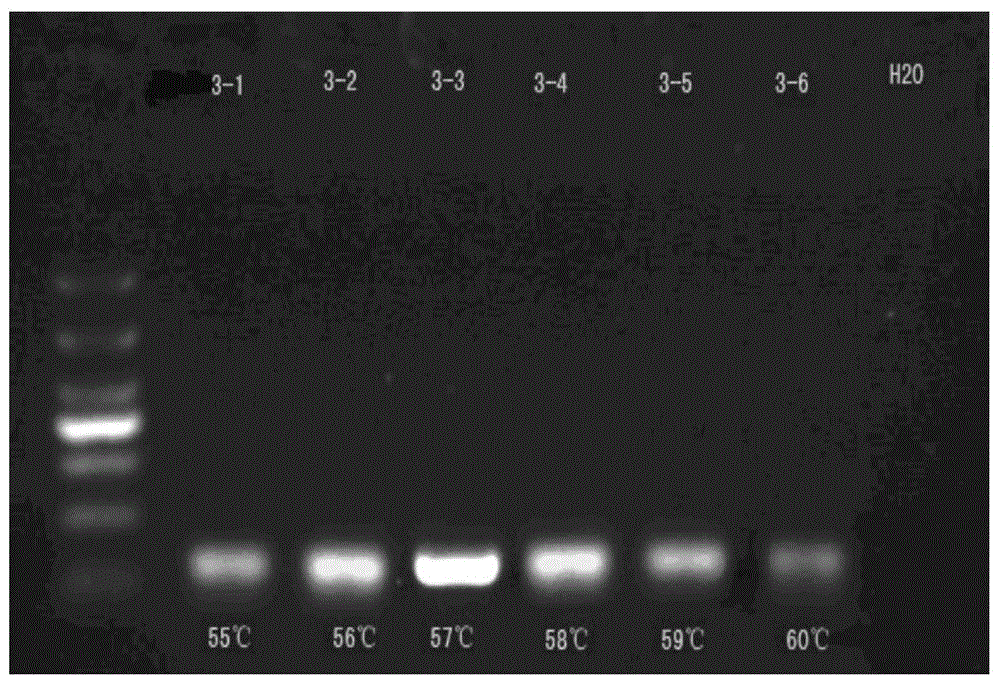
[0114] Therefore, with reference to the above-mentioned effects, the primer fragments with a size of 18-23 bp were selected for design, and the primers were synthesized by Shanghai Sangon Biotechnology Co., Ltd. The primers were verified by agarose gel electrophoresis, and the preferred forward and reverse primers were obtained, as shown in the above table. figure 1 It is the electrophoresis graph obtained by...
Embodiment 2
[0115] Determination of the PCR reaction system of embodiment 2CYP3A4*1G
[0116] The PCR reaction includes PCR buffer, dNTPs, Mgcl 2 , DNA polymerase, primers, template DNA and fluorescent dye, the PCR reaction system is (total volume is 20 μ L):
[0117]
Embodiment 3
[0118] The determination of the HRM condition of embodiment 3CYP3A4*1G
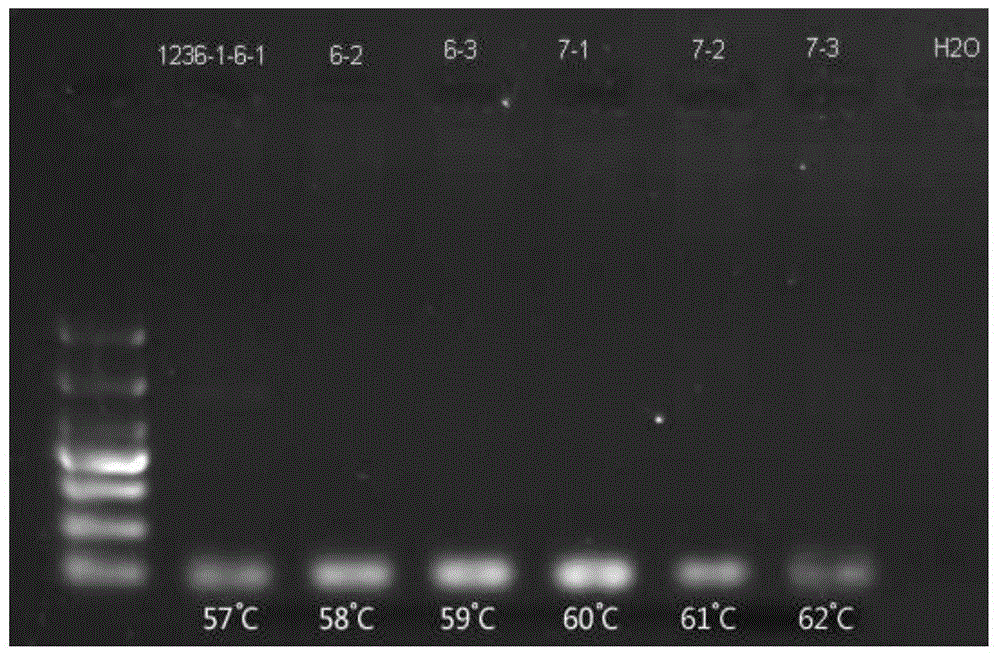
[0119] The optimization of the HRM conditions is mainly optimized and adjusted in terms of the initial temperature of the melting temperature and the number of times the fluorescence is monitored per second.
[0120] Optimal conditions:
[0121]
[0122] The PCR and HRM reaction conditions are:
[0123]
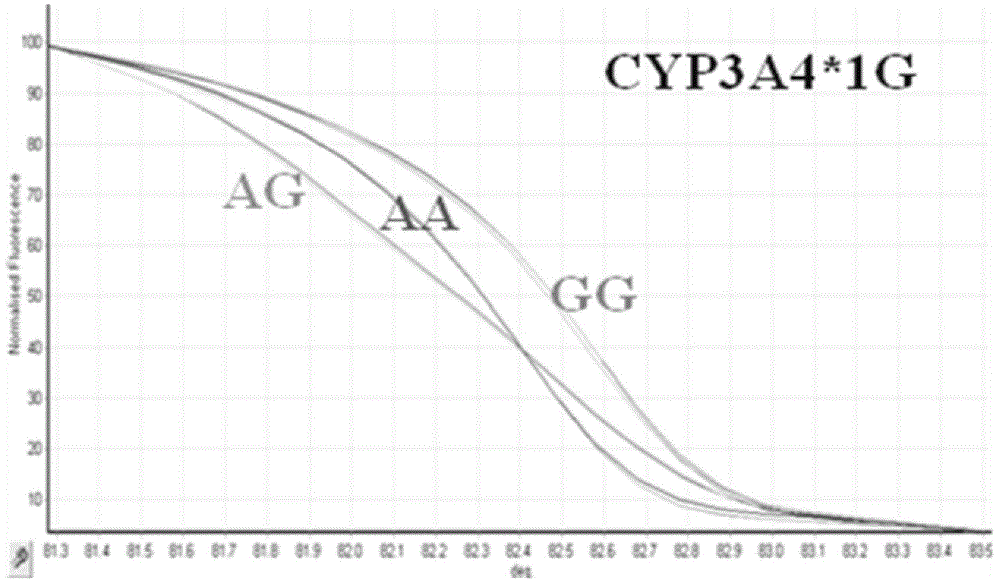
[0124] image 3 It is a high-resolution melting curve diagram of the CYP3A4*1G locus control genotype of the embodiment of the present invention;
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com