Waxberry EST-SSR molecular markers and application thereof
A technology of labeling and bayberry, applied in the direction of DNA/RNA fragments, microbial determination/inspection, recombinant DNA technology, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1: Genomic DNA was extracted by CTAB method.
[0019] (1) The formula for preparing CTAB buffer solution (hexadecyltriethylammonium bromide, Hexadecyltrimethylammonium bromide) is: 2% CTAB, 0.1MTris, 20mMEDTA, 1.4MNaCl, pH value 8.0.TE buffer solution (10mMTris, 1mMEDTA, pH8 .0).
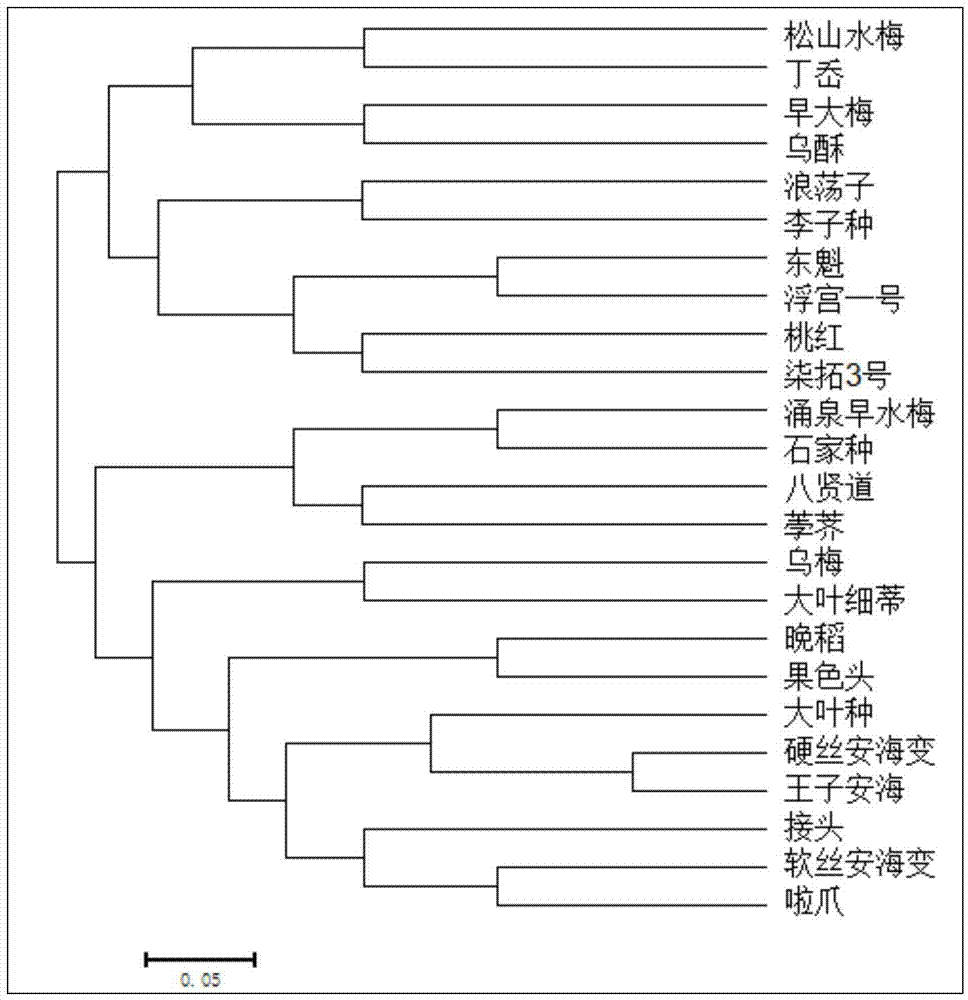
[0020] (2) Harvesting the young and tender leaves of bayberry, the variety information of bayberry is as follows figure 1 shown.
[0021] (3) Put it into liquid nitrogen and grind it to powder form, weigh about 0.7g and transfer it into a 10ml centrifuge tube filled with 4ml CTAB solution and 80μl β-mercaptoethanol (preheated at 65°C), bathe in 65°C water for 1h; add 4ml chloroform / isoamyl alcohol (24:1, v / v), mix well, centrifuge at 12000rpm for 10min, take the supernatant and add 4ml chloroform / isoamylalcohol (24:1, v / v), mix well, and centrifuge at 12000rpm for 10min . Aspirate the supernatant, add 2ml of 5M NaCl, 4ml of isopropanol (pre-cooled at -20°C), mix well, and place a...
Embodiment 2
[0022] Embodiment 2: Design EST-SSR primers
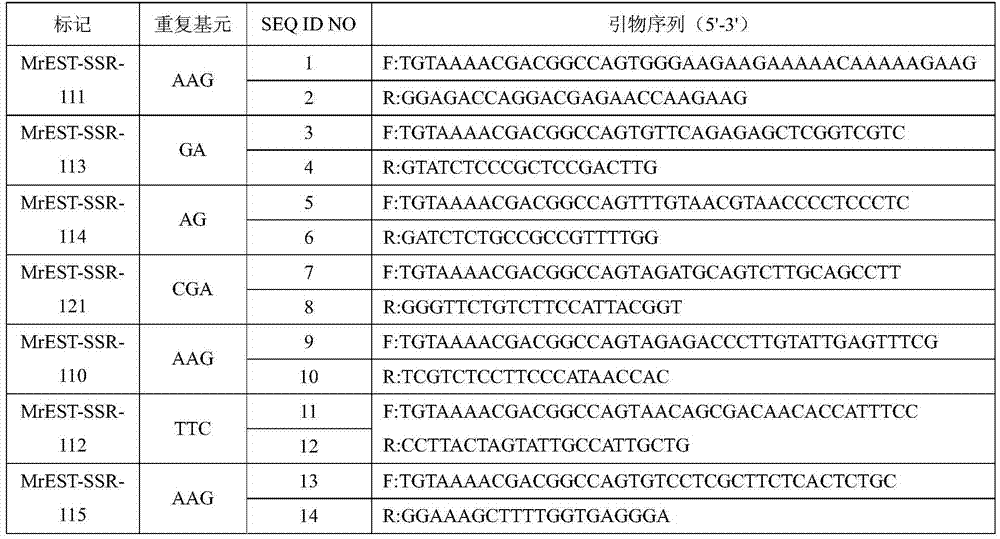
[0023] The invention obtains the EST sequence of Myrica rubra through RNA-Seq and develops EST-SSR primers. The primer design software is: NCBIPrimer-BLAST (http: / / www.ncbi.nlm.nih.gov / tools / primer-blast / index.cgi?LINK_LOC=BlastHomeAd). The specific conditions for primer design are: PCR product length 100-350bp; primer length 18-25bp; avoid primer dimer. In primer synthesis, 18 bp of M13-tail (sequence TGTAAAACGACGGCCAGT) was added to the 5' end of the forward primer. A total of 17 pairs of EST-SSR primers were developed, and the sequences are shown in Table 2.
[0024] Table 2 Marker characteristics of bayberry EST-SSR
[0025]
[0026]
Embodiment 3
[0027] Example 3: Polymorphic Primer Screening
[0028] Use the bayberry leaf DNA extracted in Example 1 as a template, and use the 17 pairs of primers designed in Example 2 to amplify.
[0029] 1. PCR reaction system: 10×ExTaqBuffer (containing Mg 2+ ), 1.6 μl of 2.5mMdNTP, 5units / μl ExTaqDNApolymerase0.1μl, forward primer 2pmol, reverse primer 8pmol, M13 universal fluorescent primer 6pmol, 10ng / μl DNA template 1μl, ddH 2 O Supplement the system to 20 μl.
[0030] 2. Use Eppendorf Mastercycler (Eppendorf Scientific, Inc.) PCR instrument to amplify. The reaction program is: 94°C pre-denaturation for 5min, then 94°C (30s) / 60°C (30s) / 72°C (30s) cycle 20 times, then 94°C (30s) / 55°C (30s) / 72°C (30s) ) cycled 12 times, and finally extended at 72°C for 15min.
[0031] 3. Electrophoresis detection: Take 5 μl of PCR products and add 1 μl of 6×loading buffer, detect on agarose gel with a concentration of 1%, and take pictures for records.
[0032] 4. STR fragment length analysis: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com