Method for performing multi-target-site amplification library construction on plasma free DNA (deoxyribonucleic acid)
A multi-target site and plasma technology, applied in the field of multi-site amplification library for plasma free DNA or short fragment DNA, to avoid primer complementation and mismatch, reduce initial requirements, and simplify primer design
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
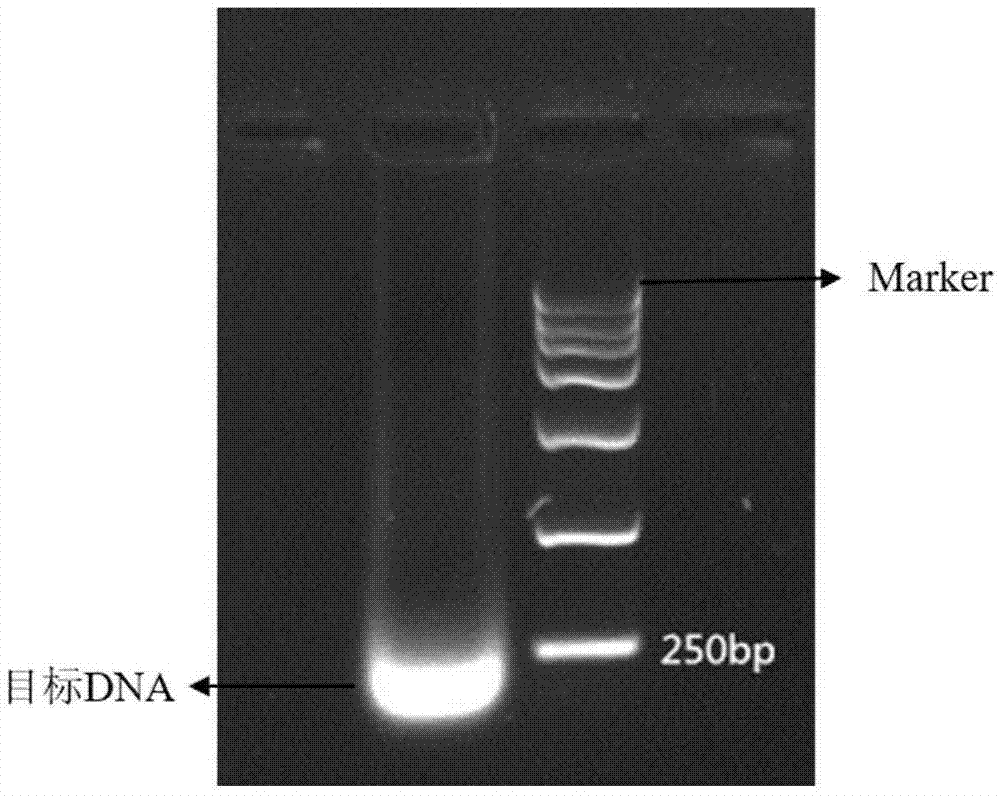
Examples
Embodiment 1
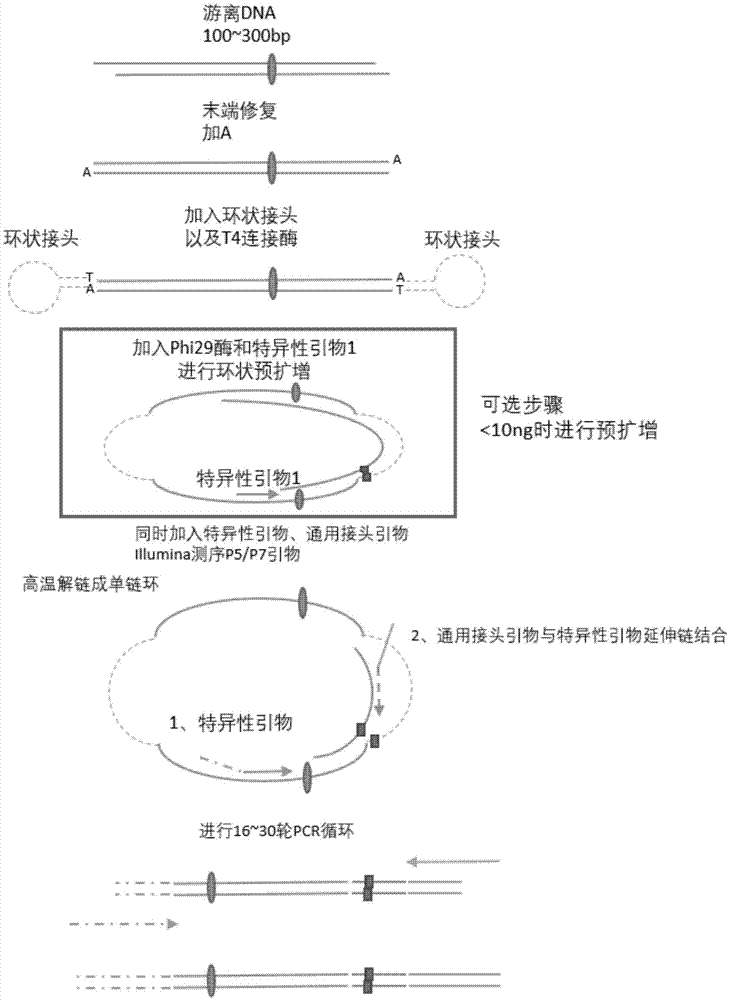
[0048] Embodiment 1, carry out multi-target site amplification method for building a library of plasma cell-free DNA, the following steps are carried out in sequence:
[0049] 1), end repair and add A
[0050] Prepare the reaction mixture according to the ratio in Table 1 below, and use a gun to gently blow up and down to mix evenly. Ultra TM IIEndRepair / dA-TailingModule (E7546) includes two parts: NEBNextUltraIIEndPrepEnzymeMix and NEBNextUltraIIEndPrepReactionBuffer, and the initial amount of free DNA is in the range of 500pg-lug (for example, 10ng).
[0051] Table 1
[0052] NEBNext Ultra II End Prep Enzyme Mix
3ul
NEBNext Ultra II End Prep Reaction Buffer
7ul
Cell-free DNA obtained from the above preparatory steps
50ul
Total
60ul
[0053] Place it in the PCR instrument and set the program reaction:
[0054] Set the top cover temperature to ≥75°C
[0055] 20℃30min
[0056] 65℃30min
[0057] Store at 4°C;
[0058] A ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com