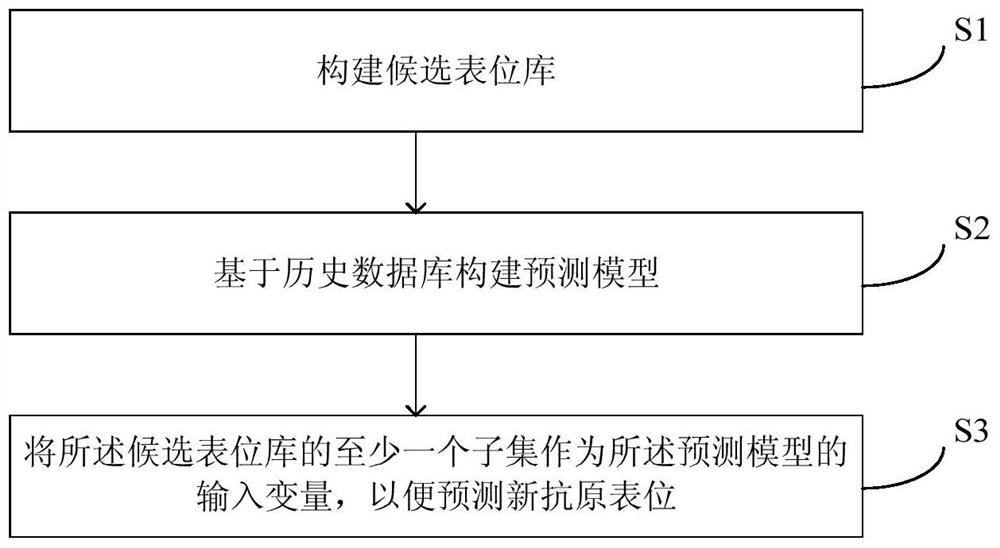
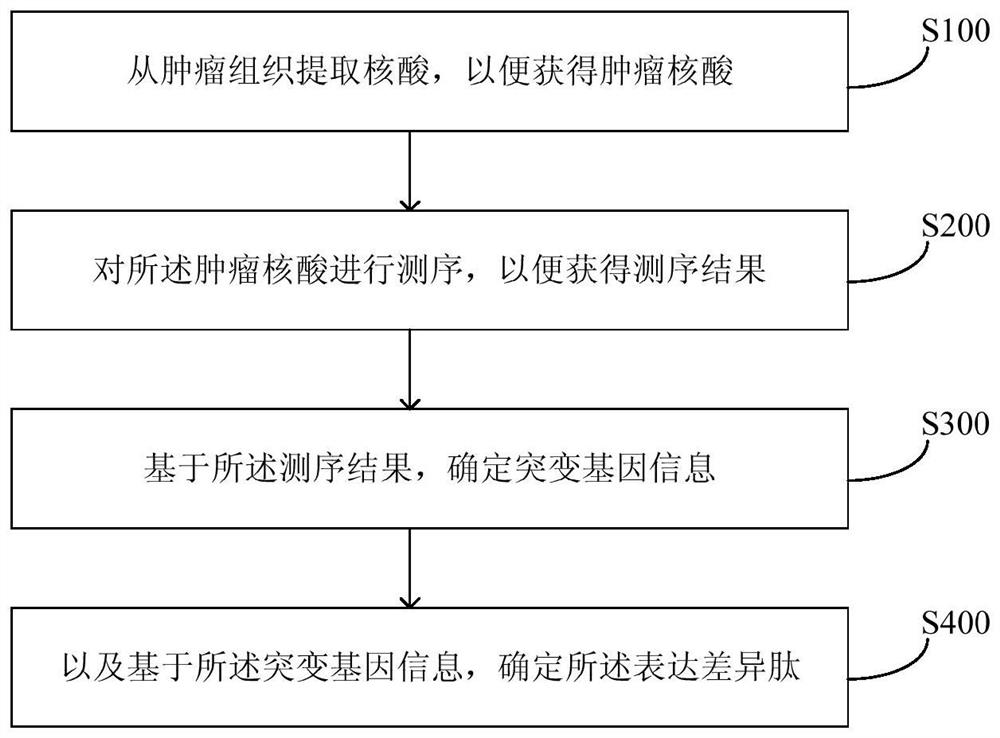
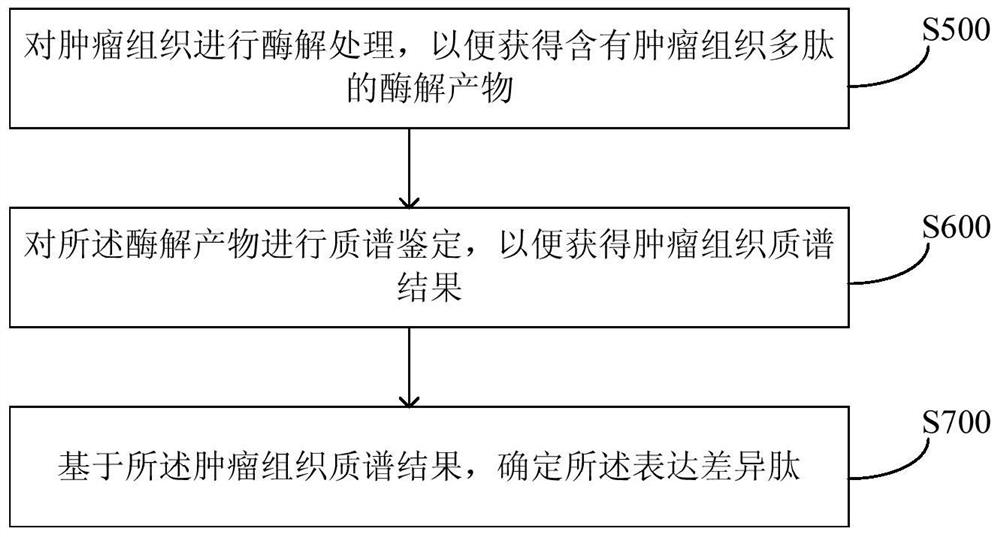
Method and equipment for predicting neoantigen epitopes
An antigen epitope and prediction model technology, applied in the biological field, can solve the problem that antigen selection needs to be deepened, and achieve the effect of good quality, high reliability and high sequencing throughput of sequencing results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0120] Illumina HiSeq 2000 was used to perform exome sequencing on transplanted tumors and peripheral blood of C57 mouse liver cancer. The tumor group sequencing data is 6.12GB, and the peripheral blood sequencing data is 6.79GB. In the first step, the short sequence alignment software BWA was used to compare the tumor group sequencing data and peripheral blood sequencing data to the mouse reference genome sequence mm9. Step 2: Use the single nucleotide polymorphism (SNP) detection software SOAPsnp to compare the results of tumor sequencing data and peripheral blood sequencing data to detect SNP, and then use the peripheral blood sample as a control to screen out the tumor data. List of somatic single nucleotide non-synonymous mutation sites. The third step is to extract a characteristic mutant peptide with a length of 10 according to the list of single nucleotide non-synonymous mutation sites. Specificity means that the peptide does not exist in the database (Ensembl release...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com