Specific SNP molecular marker for identifying cabbage clubroot 4# physiological race resistance and application
A technology for Chinese cabbage clubroot and Chinese cabbage, which is applied to the determination/inspection of microorganisms, recombinant DNA technology, biochemical equipment and methods, etc., can solve problems such as unsatisfactory control effects, achieve accurate and reliable identification results, and speed up Chinese cabbage. Effect of anti-clubroot breeding process and labor cost reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Example 1, the acquisition of molecular markers
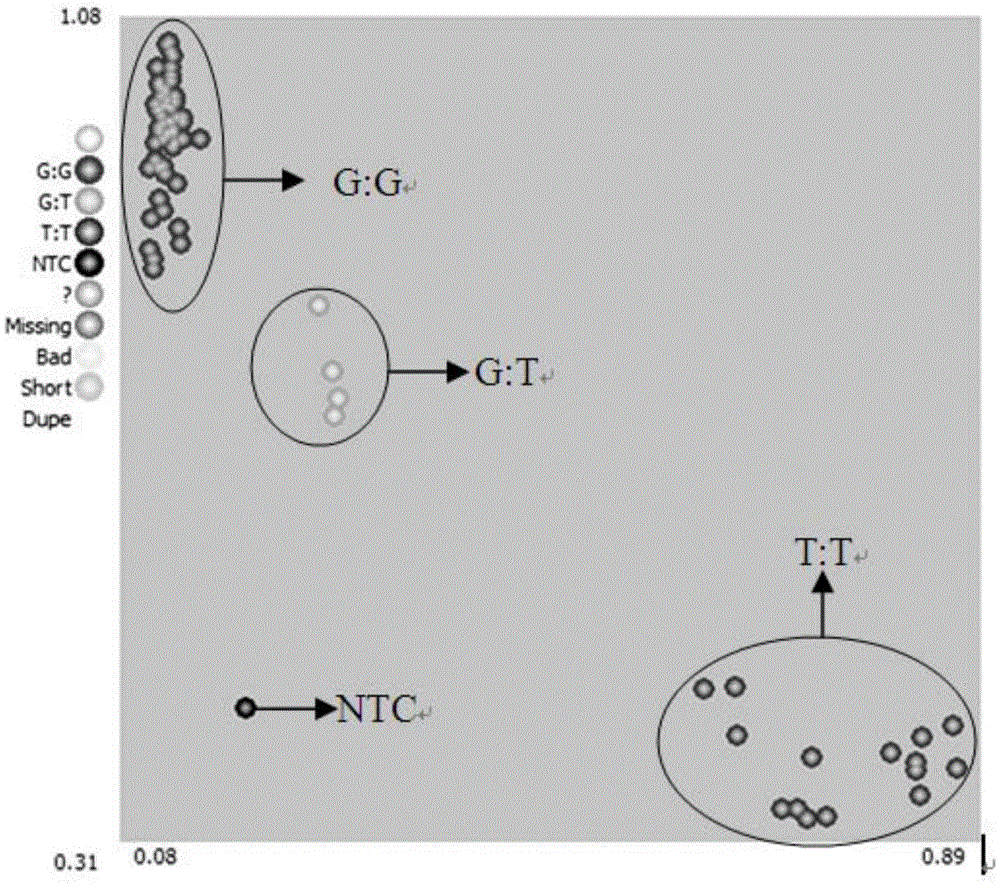
[0075] In order to screen and obtain clubroot resistance resources better and faster, and to promote the practice of molecular marker-assisted selection breeding, through the QTL mapping of the DH population composed of 68 lines combined with the clubroot phenotype, obtained on the A02 chromosome A major QTL closely linked to clubroot resistance was identified. Further design and screen SNP markers in this interval, and finally find that there is a SNP site with a G / T mutation at base 20509373 on chromosome A2 (nucleotide 301 in Sequence Listing 4), the SNP site Named A0220509373G / T site. According to the A0220509373G / TSNP site, the following specific primers that can be used in KASP technology were designed as molecular markers: upstream primer A0220509373-FF, upstream primer A0220509373-FV and downstream primer A0220509373-R.
[0076] Above-mentioned A0220509373-FF, A0220509373-FV and A0220509373-R primers were entru...
Embodiment 2
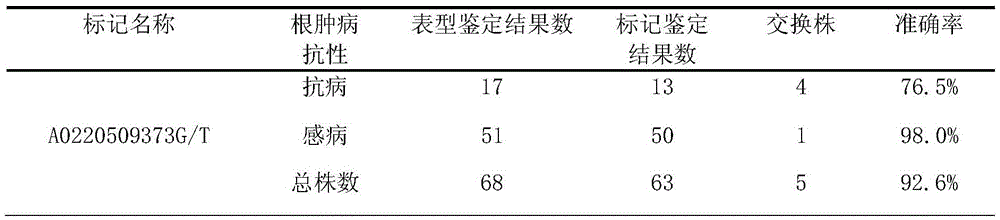
[0085] Example 2, Application of A0220509373 Molecular Marker in Identification of Chinese Cabbage Clubroot No. 4 Physiological Race Resistance
[0086] 1. Molecular marker identification of Chinese cabbage clubroot and Chinese cabbage clubroot materials
[0087] 1) DNA extraction
[0088] The genomic DNA of the 68 DH populations in Table 1 was extracted by conventional CTAB method.
[0089] The quality of the extracted DNA was detected by agarose electrophoresis and Nanodrop2100 respectively, and it was found that the extracted genomic DNA met the relevant quality requirements, that is, the agarose electrophoresis showed a single DNA band without obvious dispersion; the A260 / 280 detected by Nanodrop2100 was between 1.8- Between 2.0 (DNA sample without protein contamination); A260 / 230 between 1.8-2.0 (DNA sample with low salt ion concentration); no obvious light absorption at 270nm (DNA sample without phenol contamination); for competitive allele The amount of DNA detected b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com