A polymerase chain displacement reaction detection method for citrus huanglongbing bacteria asiatic species
A technology of citrus Huanglongbing and substitution reaction is applied in the field of polymerase chain displacement reaction detection of citrus Huanglongbing Asian species, which can solve the problems of difficult separation and identification of amplified products, and achieves the improvement of molecular diagnosis accuracy and false positives. Effects with low probability and broad market prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0034] The principles and features of the present invention are described below in conjunction with the accompanying drawings, and the examples given are only used to explain the present invention, and are not intended to limit the scope of the present invention.
[0035] 1. Principle explanation
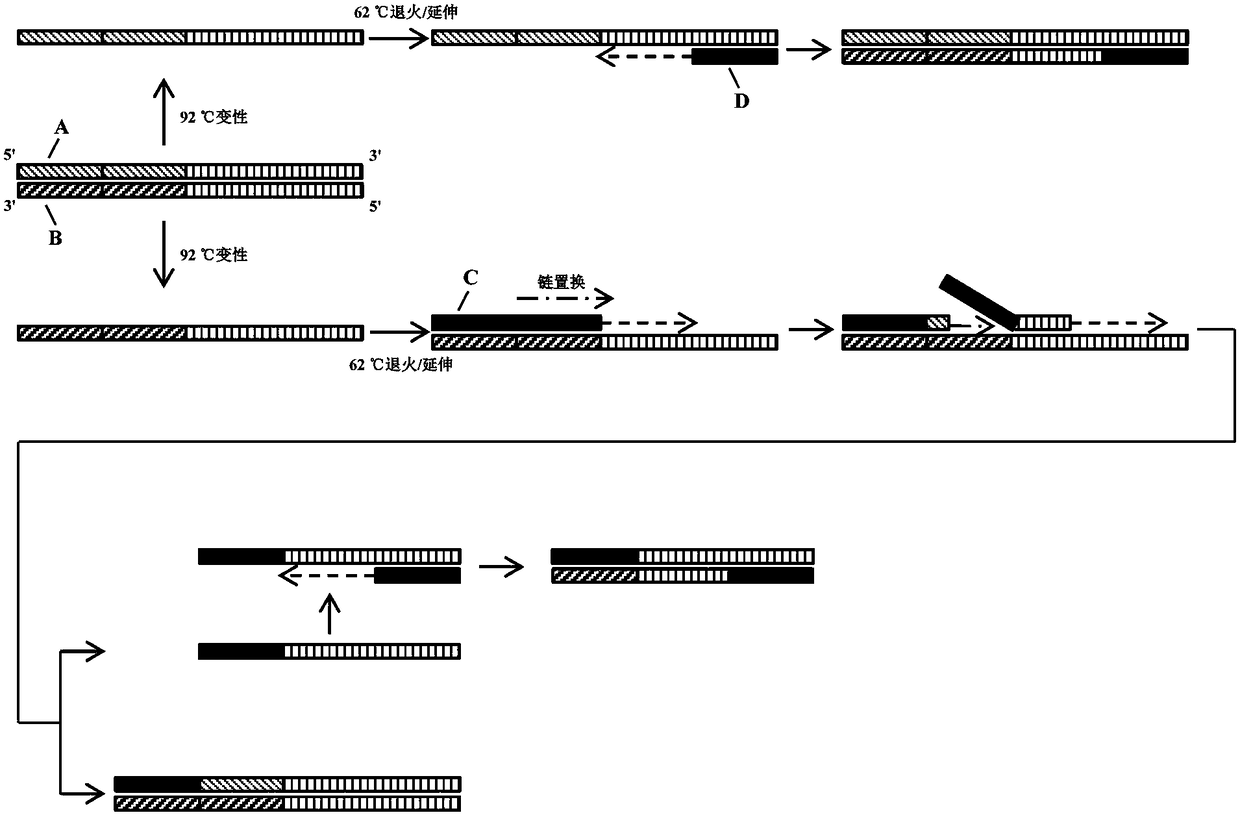
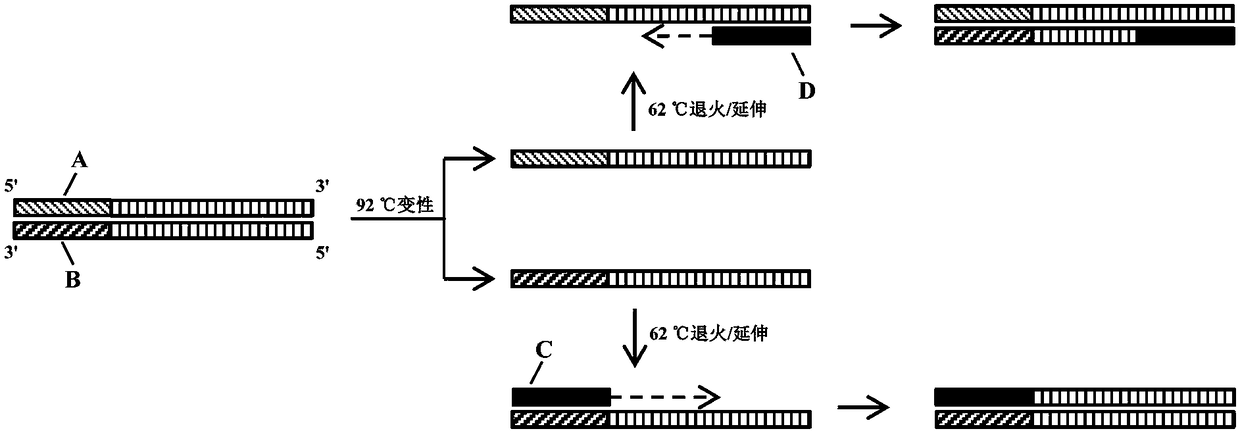
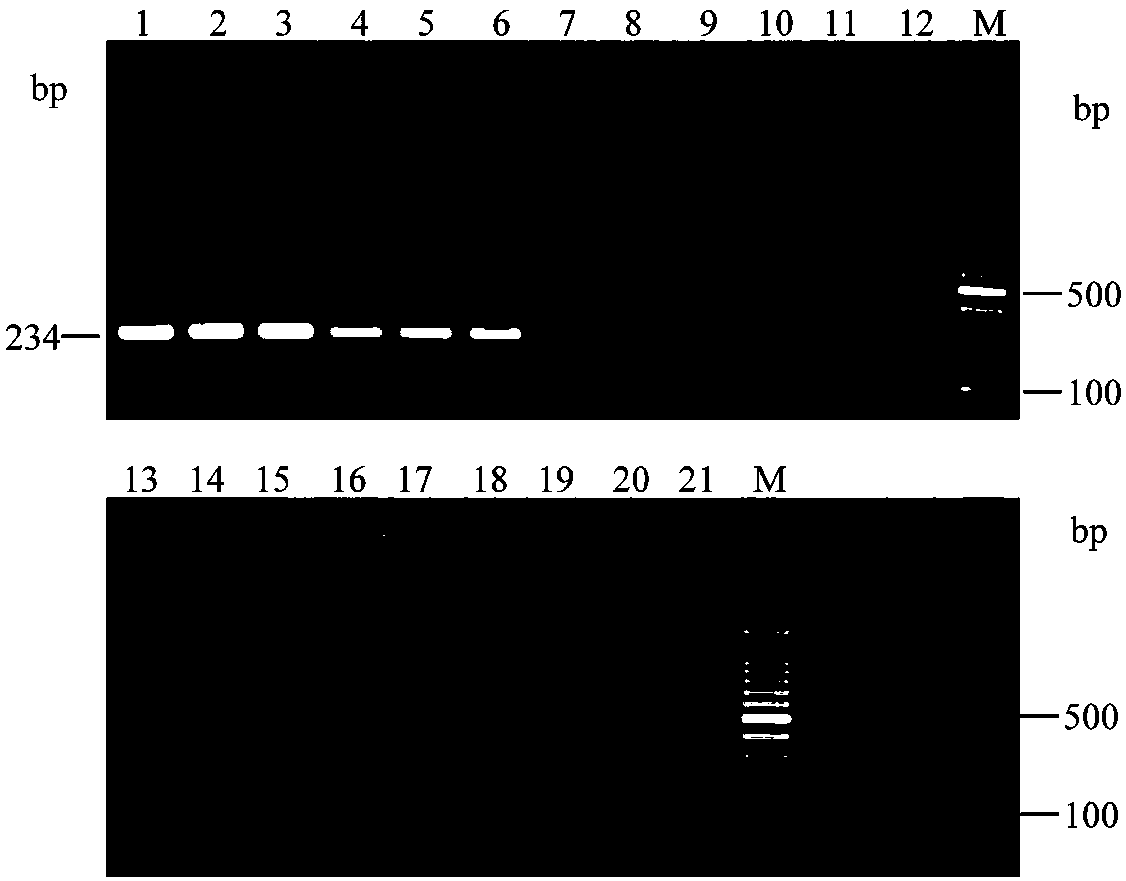
[0036] Such as figure 1 As shown, on the genome of H. citriensis Asiatic species, there are two upstream primer binding sites, which are two tandem repeat sequence units, and only one downstream primer binding site, which is a sequence-specific nucleic acid region. Since the DNA polymerase (SD enzyme) used in the reaction has dual characteristics of heat resistance and strand displacement activity, in the first round of reaction, the target nucleic acid fragment (double-stranded DNA) on the genome of Huanglongbing citri Denatured, unzipped into two DNA single strands, and then at 62°C annealing / extension temperature: the two upstream primers simultaneously bind to the DNA single-st...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com