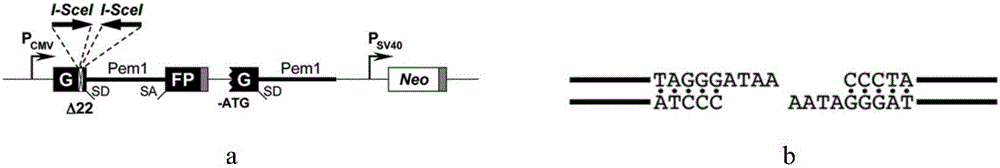Method for increasing HR (Homologous Recombination) repairing frequency of ovine embryo fibroblasts after gene editing
A fibroblast and gene editing technology, applied in the biological field, can solve the problems of low probability of homologous recombination and low efficiency of gene targeting
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1. Screening of siRNAs Interfering with Lig4 Gene Expression in Sheep Embryo Fibroblasts
[0042] 1. Construction of eukaryotic expression plasmid pcDNA3.1-Shlig4
[0043] 1. Primer design
[0044] Primers were designed according to the predicted sheep Lig4 gene sequence (XM_004012236) and GAPDH gene sequence (NM_001289746.1) in Genbank, and were synthesized by Shanghai Sangon Biotechnology Co., Ltd. The primer sequences are listed in Table 1.
[0045] Table 1 is the primer name and sequence
[0046]
[0047] The underlines are the restriction sites BamHI and XbaI, and the boldfaces are the protective bases.
[0048] 2. Construction of pcDNA3.1-Shlig4 recombinant plasmid
[0049] According to the cDNA sequence of the mammalian Lig4 gene without introns, use the primers Shlig4F / Shlig4R to directly use the sheep genomic DNA as a template for PCR amplification, and obtain a 2736bp PCR product that matches the expected size, which is the cDNA of the sheep Lig...
Embodiment 2
[0074] Example 2, Improving the frequency of homologous recombination repair of sheep embryonic fibroblast genomic DNA
[0075] Lig4 gene siRNA improves HR repair efficiency in isolated sheep embryonic fibroblasts, as follows:
[0076] Due to the limited passage capacity of sheep primary fibroblasts, it is basically not feasible to establish stable clones. For quantitative determination of DNA end joining efficiency and fidelity, a plasmid rejoining assay (plasmid rejoining assay) was used.
[0077] The HR plasmid donated by the Gorbunova laboratory uses GFP (Green Fluorescent Protein) as a refactored reporter gene. The reporter system contains 2 copies of mutated GFP-Pem1. In the first GFP-Pem1 gene fragment, the first exon of GFP contained a deletion of 22 bp base length, and two reverse symmetric I-SceI enzyme recognition sites were inserted. The second GFP-Pem1 gene fragment lacks the promoter, ATG start codon and exons of the second part of GFP. When the I-SceI site i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com