Enzyme composition for dna end repair, adenylation, phosphorylation
An enzyme composition and end repair technology, applied in the field of enzyme compositions, can solve problems such as damage to genome sequence assembly, complex genome sequence assembly, damage to genome assembly and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Prove that different enzymes require different buffers
[0044] It is known in the art that each enzyme has different optimal storage and reaction conditions. Enzyme storage conditions are often different from the recommended reaction conditions shown in Table 2. Table 2 shows the storage and reaction buffer components of DNA inactivation, phosphorylation, and dA tailing enzymes sold by the following commercial suppliers: Thermo Fisher Scientific, New England Biolabs and Life Technologies.
[0045] Table 2. Storage buffer (SB) and reaction buffer (RB) of enzymes used for passivation, phosphorylation and dA tailing of DNA fragments
[0046]
[0047]
[0048]
[0049] In order to obtain the composition of the present invention, all the above-indicated enzymes T4 DNA polymerase, Klenow fragment, T4 DNA polynucleotide kinase and modified thermophilic polymerases with end-tailing activity, such as modified Brucella The thermobacterium or Thermus aquaticus DNA polymerase must be p...
Embodiment 2
[0054] Effect of buffer and polymerase composition on passivation, tailing and phosphorylation
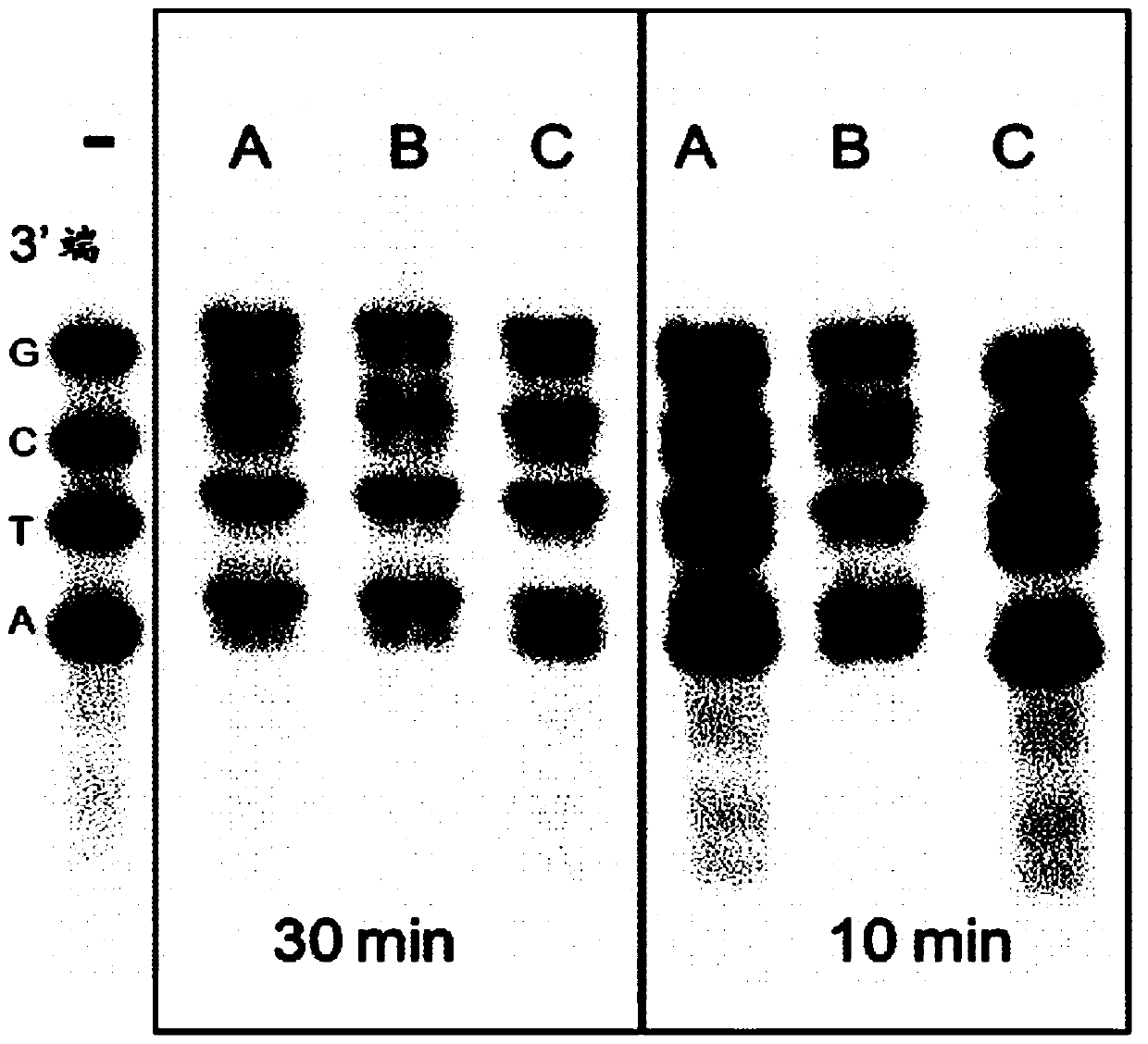
[0055] To test the efficiency of 3'end tailing of DNA fragments and analyze the potential bias of the 3'end nucleotides, we developed and used a kind of different length and 3'end nucleotides, and used it at the 5'end Cy5 labeled four oligonucleotide duplex model system ( figure 1 ).
[0056] Check Klenow fragment exo - The mutant’s dA tailing ability and the figure 2 It is shown that lane (A) is the best Klenow fragment buffer (10x reaction buffer, EP0421) supplemented with 0.2mM dATP; lane (B) is commercial buffer G containing 1mM DTT and 0.2mM dATP; and lane ( C) is the optimized DNA end repair buffer (fast DNA end repair kit K0771). Use 5 units of enzyme in a 50μl reaction mixture (containing as figure 1 A dA tailing reaction was performed in the shown Cy-5 labeled oligonucleotide duplex (7.5 pmol equivalent molecular mixture). The pair of bands represents the oligonucleotide d...
Embodiment 3
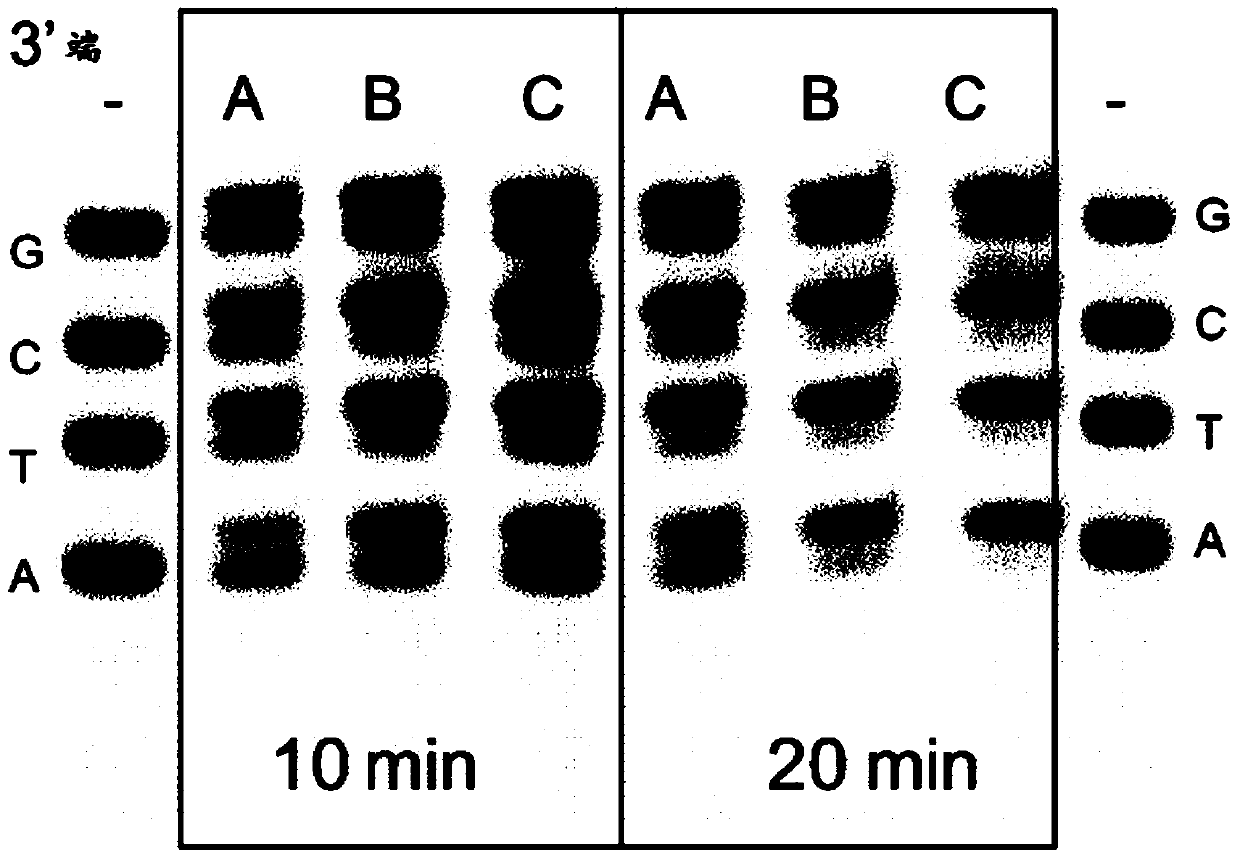
[0071] Stability of 2x concentrated terminal transformation master mix
[0072] For stability testing, the 2x terminal transformation master mix as disclosed was stored at -20°C and +25°C (to represent accelerated stability testing) for different periods of time. use for Figure 5 Perform stability testing with the same experimental protocol shown in. Incubate 1 μg control DNA fragment in 50 μl reaction mixture of 1x end transformation master mix that has been stored at -20°C for 5 min to allow end repair enzymes to inactivate and phosphorylate the DNA ends, and then incubate at 72°C for 10 minutes. Conducive to the simultaneous inactivation of the mesophilic DNA end repair enzyme and the 3'-end tailing of the DNA by mod-Tbr DNA polymerase. After this step, a DNA adaptor with a length of 60 bp carrying a 3'terminal dT extension was added to the reaction mixture to a final concentration of 1 μM, and ligation was performed at 22° C. for 5 min using T4 DNA ligase. The resulting re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com