A new method for label-free detection of t4 polynucleotide kinase/phosphatase and its inhibitors based on fluorescent copper nanoparticles
A technology of polynucleotides and nanoparticles, applied in biochemical equipment and methods, microbiological determination/inspection, etc., can solve the problems of low selectivity and sensitivity, high cost, application limitations, etc., and achieve good selectivity , cost reduction, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
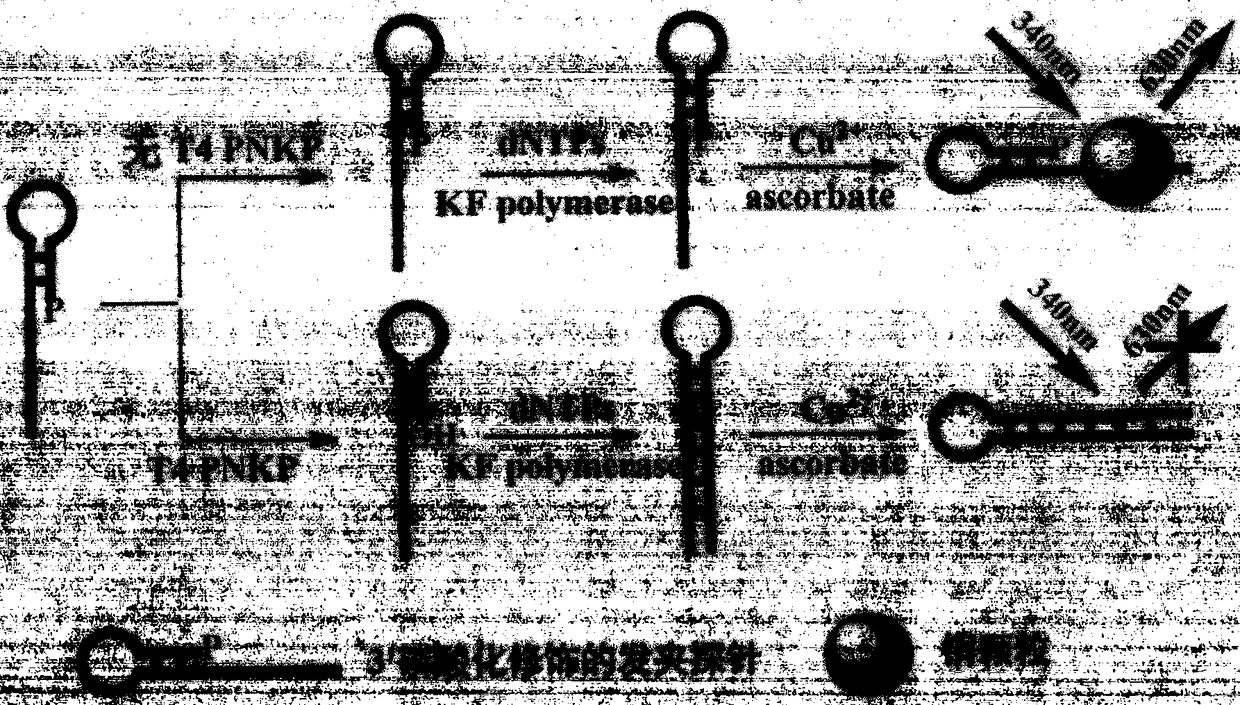
[0040] Step 1. Design the probe sequence as follows: 5′-TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT CGC ACC TAA AG G GTG CG -3' (phosphorylation modification at the 3' end);
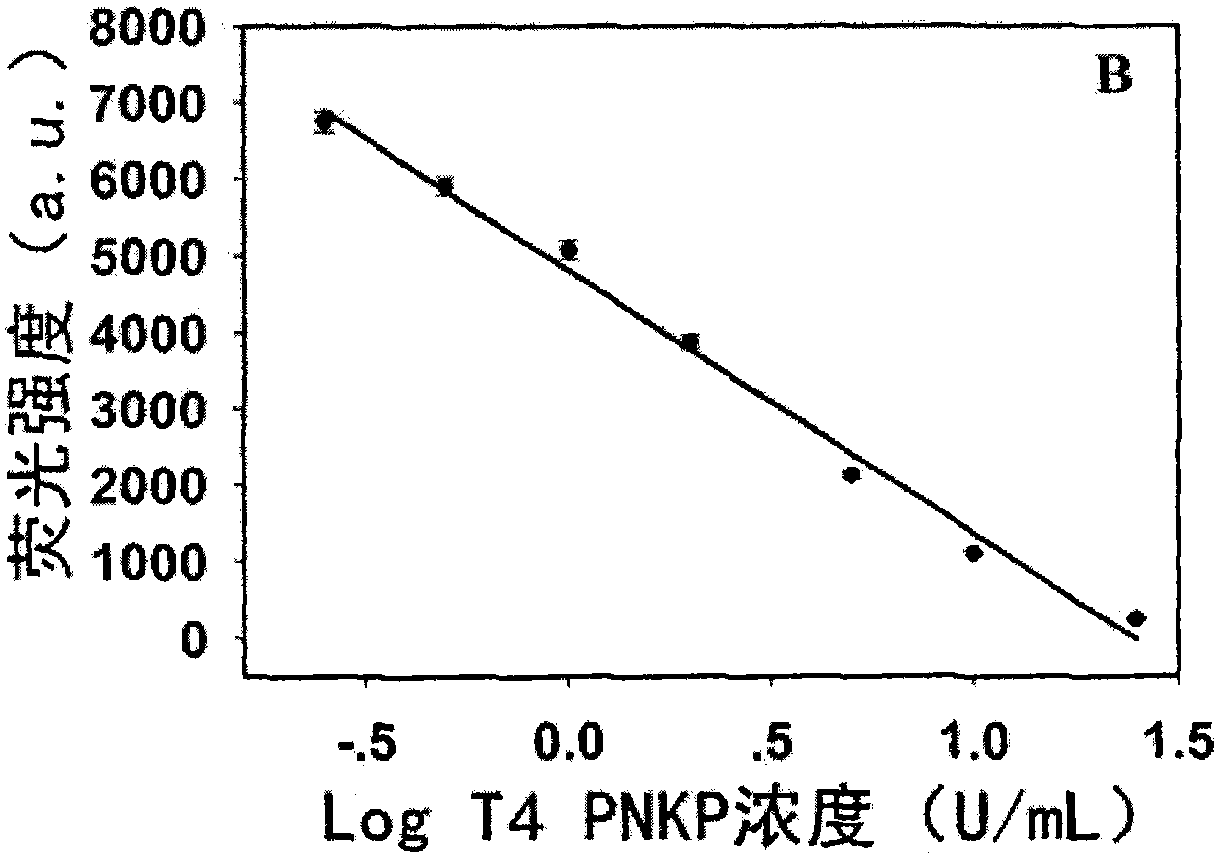
[0041] Step 2. 25 μL Tris-HCl buffer system (10 mM Tris-HCl, 10 mM MgCl, 50 mM NaCl, pH7.9) contains 5 μL probe (1 μM), 1 μL KF polymerase (50 U / mL), 2 μL dNTPs (200 μM) and 2 μL of different concentrations of T4PNKP (0U / mL, 0.25U / mL, 0.5U / mL, 1U / mL, 2U / mL, 5U / mL, 10U / mL, 25U / mL), mixed well, and kept at 37℃ Incubate for 80min;
[0042] Step 3. Supplement the above solution into MOPS buffer solution (10 mM MOPS, 150 mM NaCl, pH 7.6) with a final volume of 100 μL, including 10 μL sodium ascorbate (2 mM) and 10 μL copper sulfate (200 μM), mix well, and incubate at room temperature for 10 min ;
[0043] Step 4, the final reaction solution obtains the fluorescence spectrum figure of 520~660nm under the excitation wavelength of 340nm (see Figure 2a ) and the linear correspondence between the fluore...
Embodiment 2
[0045]Step 1. Design the probe sequence as follows: 5′-TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT CGC ACC TAA AG G GTG CG -3' (phosphorylation modification at the 3' end);
[0046] Step 2, 25 μL Tris-HCl buffer system (10 mM Tris-HCl, 10 mM MgCl, 50 mM NaCl, pH7.9) contains 5 μL probe (1 μM), 1 μL KF polymerase (50 U / mL), 2 μL dNTPs (200 μM), 5 μL of different concentrations of heparin (0 μg / mL, 0.2 μg / mL, 0.5 μg / mL, 1 μg / mL, 2 μg / mL, 5 μg / mL, 10 μg / mL, 25 μg / mL) and 2 μL of T4PNKP (25 U / mL), mixed Evenly, incubate at 37°C for 80 minutes;
[0047] Step 3. Supplement the above solution into MOPS buffer solution (10 mM MOPS, 150 mM NaCl, pH 7.6) with a final volume of 100 μL, including 10 μL sodium ascorbate (2 mM) and 10 μL copper sulfate (200 μM), mix well, and incubate at room temperature for 10 min ;
[0048] Step 4, the corresponding relationship between the relative fluorescence intensity of the system and the concentration of heparin is obtained from the final rea...
Embodiment 3
[0050] Step 1. Design the probe sequence as follows: 5′-TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT CGC ACC TAA AG G GTG CG -3' (phosphorylation modification at the 3' end);
[0051] Step 2, 25 μL 1% lung cancer cell lysate Tris-HCl buffer system (10mM Tris-HCl, 10mM MgCl, 50mM NaCl, pH 7.9) contains 5 μL probe (1 μM), 1 μL KF polymerase (50 U / mL), 2 μL dNTPs (200μM) and 2μL of different concentrations of T4 PNKP (0U / mL, 0.25U / mL, 0.5U / mL, 1U / mL, 2U / mL, 5U / mL, 10U / mL, 25U / mL), mix well, in Incubate at 37°C for 80 minutes;
[0052] Step 3. Supplement the above solution into MOPS buffer solution (10 mM MOPS, 150 mM NaCl, pH 7.6) with a final volume of 100 μL, including 10 μL sodium ascorbate (2 mM) and 10 μL copper sulfate (200 μM), mix well, and incubate at room temperature for 10 min ;
[0053] Step 4, the final reaction solution obtains the fluorescence spectrum figure of 520~660nm under the excitation wavelength of 340nm (see Figure 4a ) and the corresponding relati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com