Triple fluorescent quantitative PCR primer, probe and kit for identifying pseudorabies virus strains
A pseudorabies virus, fluorescent quantitative technology, applied in the field of a-K61 vaccine, can solve the problems of inability to distinguish between classic and mutant strains of PRV, time-consuming and labor-intensive problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
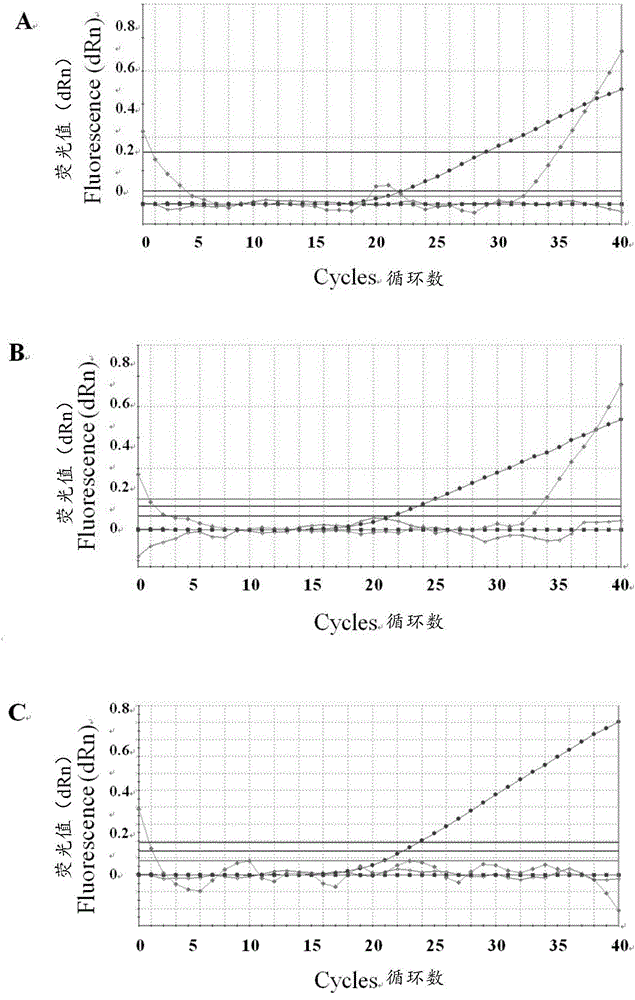
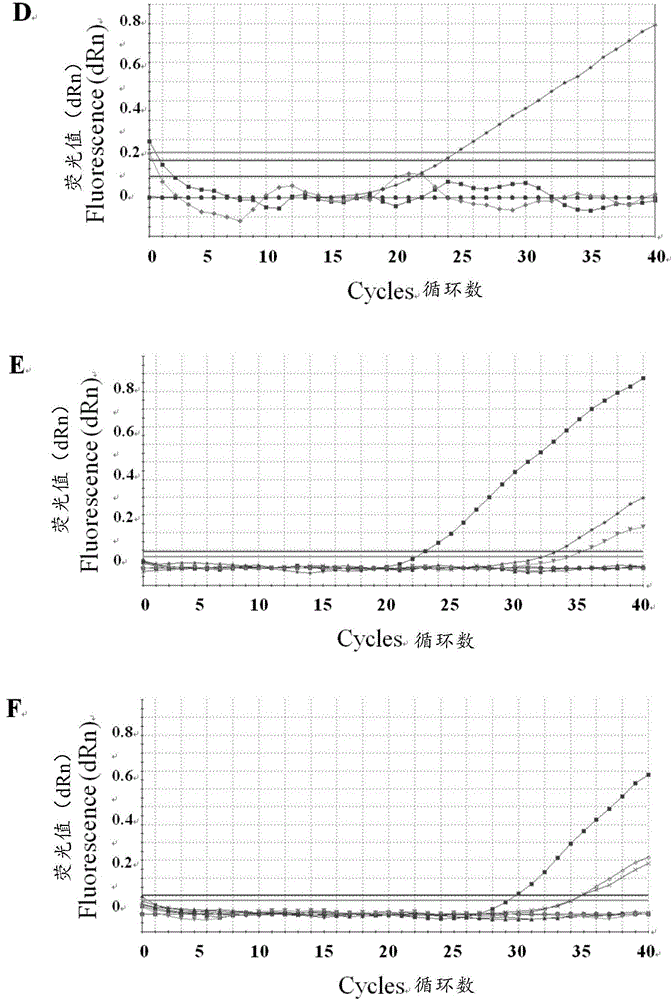
[0030] Example 1 Establishment of triple fluorescent quantitative PCR method for identification and detection of PRV classic strain, variant strain and Bartha-K61 vaccine strain
[0031] 1. Materials and methods
[0032] 1.1 Virus
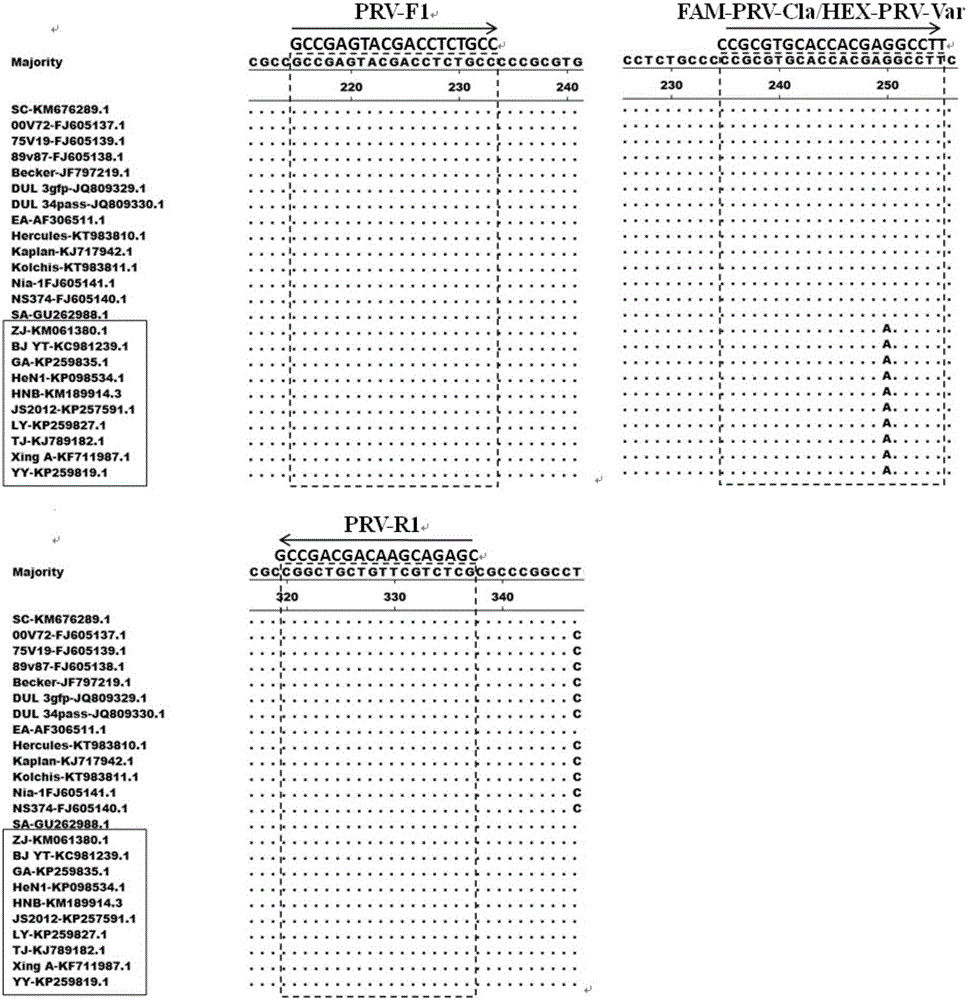
[0033] Swine fever virus (Classicalswinefevervirus, CSFV) Shimen strain, porcine parvovirus (Porcineparvovirus, PPV) Z strain, porcine circovirus type 2 (Porcinecircovirustype2, PCV2) JXL strain, porcine reproductive and respiratory syndrome virus (Porcinereproductiveandporcinerespiratorysyndromevirus, PRRSV) HuN4 strain , PRVSC strain, TJ strain, HLJMDJ2013 strain, HeBLP2014 strain, BJKJZ2015 strain (variant strain) and Bartha-K61 (vaccine strain) are preserved by the present inventor (among which the pseudorabies virus TJ strain, HLJMDJ2013 strain, HeBLP2014 strain and BJKJZ2015 strain, are by the present invention Humans were isolated, identified, and preserved, and their partial sequences were uploaded to GenBank at the same time, with sequenc...
Embodiment 2 3
[0049] Example 2 Optimization of the conditions of triple fluorescent quantitative PCR method
[0050] Based on the triple fluorescence quantitative PCR method established in Example 1, the present invention further optimizes the triple fluorescence quantitative PCR conditions.
[0051] 1. Experimental method
[0052]Triple fluorescent quantitative PCR experiments were performed using the Mx3005p instrument (Stratagene, USA), and the concentrations of primers and probes were optimized by the value of the fluorescent signal (ΔRn). Experimental conditions are as follows: PCR reaction total volume is 20 μ L, FAM-PRV-Cla (shown in SEQIDNo.1), HEX-PRV-Var (shown in SEQIDNo.4) and Cy5-PRV-Bar (shown in SEQIDNo.6) ( 10 μM) each 0.5 μ L; PRV-F1 (shown in SEQIDNo.8), PRV-R1 (shown in SEQIDNo.9), PRV-F2 (shown in SEQIDNo.14) and PRV-R2 (shown in SEQIDNo.15) ( Genomic DNA 2 μL; PremixExTaqProbeqPCR (TaKaRa, Japan) 10 μL; meanwhile, 0%, 2.5%, 5% and 10% (v / v) dimethyl sulfoxide (DMSO) w...
Embodiment 3 3
[0057] Specificity, sensitivity and repeatability test of embodiment 3 triple fluorescence quantitative PCR method
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com