Efficient bidirectional transcription/expression plasmid and application thereof in influenza virus reverse genetics
A technology of reverse genetics and viruses, applied in the direction of viruses, viruses/bacteriophages, antisense single-stranded RNA viruses, etc., can solve the problems of low virus titer, difficulty in adapting to research needs, time-consuming and labor-intensive problems, and achieve high toxin production Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1 Construction and functional identification of bidirectional transcription / expression vector
[0047] 1. Construction of bidirectional transcription / expression vector
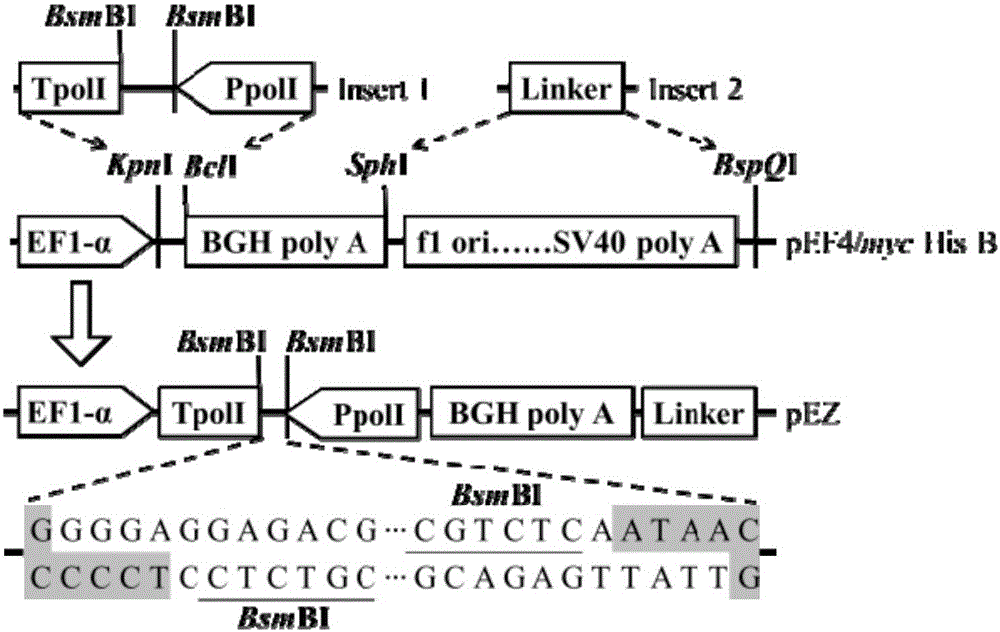
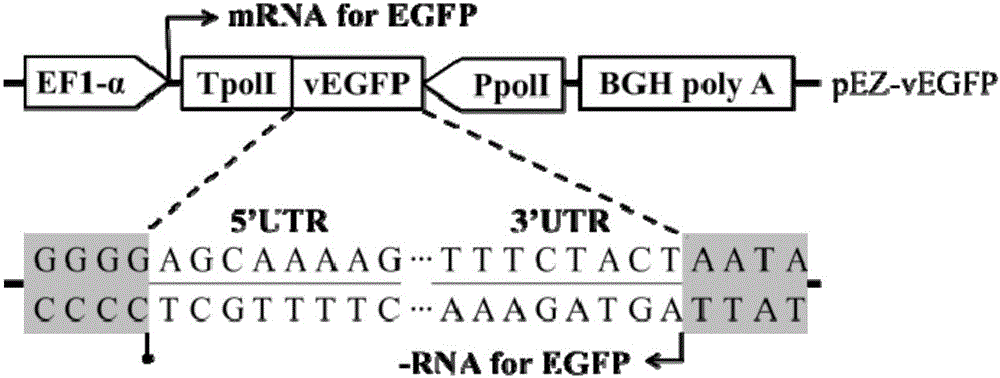
[0048] Such as figure 1 As shown, the eukaryotic expression vector pEF4 / myc-His B is used as the backbone for transformation: an essential element (Insert 1, whose sequence is shown in SEQ ID NO.11) is inserted between its KpnI and BclI restriction sites—mouse Source polI terminator (TpolI, 33bp), BsmBI restriction site and human polI promoter (PpolI, 236bp) sequence; a short Linker sequence (Insert2, Insert2, The sequence is 5'-CTGGGGATGCGGTGGGCTCTATG-3'), replacing the non-essential elements from flori to SV40poly A signal on the original vector. The transformed vector is named pEZ, and its nucleotide sequence is shown in SEQ ID NO.1.
[0049] 2. Construction of recombinant plasmid carrying reporter gene
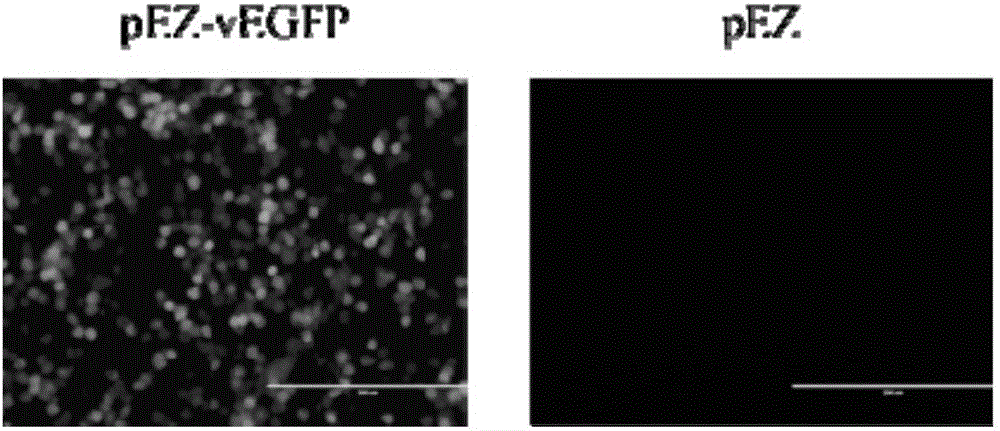
[0050] A reporter gene was inserted into the pEZ vector to verify its bidirectional ...
Embodiment 2E
[0061] The establishment of embodiment 2EIV reverse genetic system
[0062] 1. Construction of recombinant plasmid carrying viral cDNA and introduction of molecular markers
[0063] Extract the viral RNA of the EIV JL89 strain according to the instruction manual of the viral RNA extraction kit, use uni12 as the reverse transcription primer, use the corresponding primer pair of each segment (Table 1) as the amplification primer, and use the RT-PCR method to amplify 8 genes Segments (PB2, PB1, PA, HA, NP, NA, M and NS). Each segment was cleaved with the corresponding restriction endonuclease (BsmBI or BsaI), and then directional cloned into the BsmBI of pEZ, and 8 recombinant plasmids were constructed, and the 8 gene segments contained in them—PB2, PB1, The nucleotide sequences of PA, HA, NP, NA, NS and M gene segments are respectively shown in SEQ ID NO.2-9.
[0064] Table 1 Primers for amplifying 8 segments
[0065]
[0066] In addition, wild-type (wt) HA segments were n...
Embodiment 3
[0073] Identification of embodiment 3 recombinant virus
[0074] 1. Sequence determination of recombinant virus and detection of its molecular markers
[0075] The 8 segments of rJL89 were amplified according to the primers and methods described in Method 1 of Example 2, cloned into the pMD18T vector, the sequences of each segment were determined, and compared with the sequences of each segment of JL89. At the same time, the HA segment was identified by restriction endonucleases HindIII and NdeI.
[0076] result:
[0077] Eight segments of rJL89 were cloned and sequenced. The alignment of each segment of rJL89 with JL89 showed that the HA segment contained a nonsense mutation (T 449 C and C 983 T), the identity of the HA segment of JL89 is 99.9%; while the identity of PB2, PB1, PA, NP, NA, M and NS is 100% with the corresponding segment of JL89; this shows that the genetic information of rJL89 is almost entirely derived from Since JL89.
[0078]The HA segment of rJL89 co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com