Kit for simultaneously detecting multisite mutation of genes CYP2C19 and CYP2D6
A CYP2C19, multi-site mutation technology, which is applied in the direction of recombinant DNA technology, microbial measurement/inspection, biochemical equipment and methods, etc., can solve the problem of not being able to obtain multiple site information at the same time, and achieve the effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
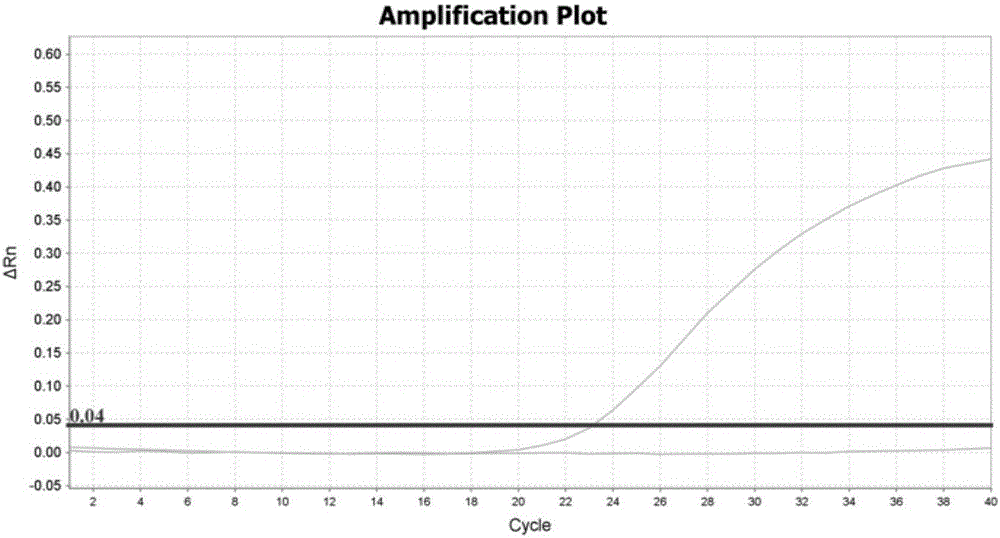
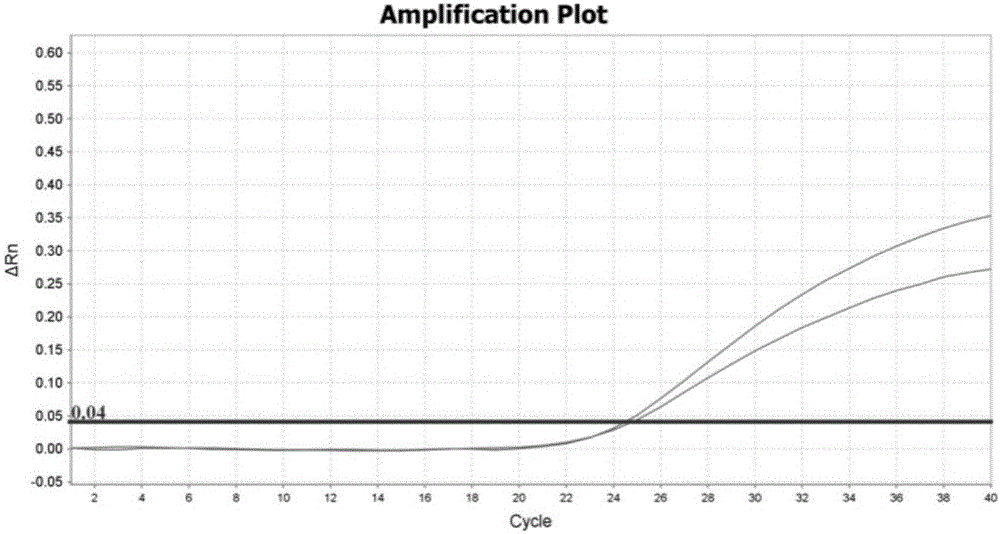
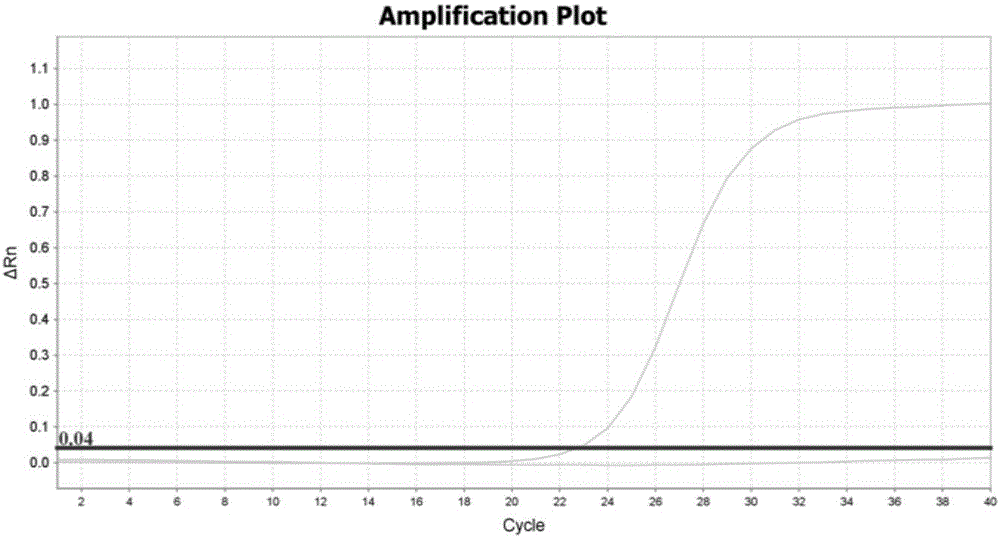
[0042] The design of embodiment 1 primer and probe
[0043] Because CYP2C19 and CYP2D6 genes have highly homologous sequences in the genome, it is generally difficult to design specific primers and probes for the Taqman allele discrimination assay for detecting CYP2D6 gene variation. The present inventors carried out careful design, repeated verification, screening and optimization, and finally obtained specific primers and probes based on the Taqman allele discrimination analysis method, as shown in Table 1. Table 2 and Table 3 are the primers and probes that cannot successfully specifically detect the mutations of CYP2C19 and CYP2D6 genes.
[0044] Table 1
[0045]
[0046]
[0047]
[0048] Table 2
[0049]
[0050]
[0051]
[0052] table 3
[0053]
[0054]
Embodiment 2
[0055] Embodiment 2 fluorescence quantitative PCR detection process
[0056] 1. A 96-well plate design with 8 rows x 12 columns is adopted, in which 6 clinical samples are detected in columns 1 to 6, and corresponding wild-type, mutant, and heterozygous quality controls are installed in columns 7 to 9. Template, columns 10 to 12 were filled with 6 different ratios of copy number standards (Table 4).
[0057] Table 4 shows an example of a suggested layout for a 96-well PCR reaction plate
[0058]
[0059]
[0060] Note: S stands for sample, S1 to S6 stands for 6 samples; W stands for wild-type quality control template; M stands for mutant quality control template; H stands for heterozygous quality control template; NTC stands for wells without DNA samples; STD stands for Copy number gradient standard; where STD1 to STD6 represent 10-fold serial dilutions of the plasmid standard containing CYP2D6 and ALB gene fragments (1:10, 1:100, 1:1000, 1:10000, 1:100000 and 1 :10000...
Embodiment 3
[0073] The assembly of embodiment 3 detection kits
[0074] A kit for simultaneously detecting multi-site mutations in CYP2C19 and CYP2D6 genes, comprising the following components: each primer described in Table 1 with a concentration of 0.25-0.75 μM and a volume of 0.5-1.25 μL (including primers designed for each gene locus) Forward primer and reverse primer, for example, for CYP2D6*2 site, add primers for detecting CYP2D6*2 site), the concentration of each probe described in Table 1 is 0.25-0.75 μM, and the volume is 0.125-0.625 μL Needle (including wild allele probes and mutant allele probes designed for each gene locus, for example, for CYP2D6*2 loci, add probes for detecting CYP2D6*2 loci), 5-15 μL 2 ×Taqman Master Mix, make up the total volume with double distilled water to 10-25 μL; wild-type quality control template; mutant quality control template; heterozygous quality control template.
[0075] In order to more concisely illustrate the composition of the above kit,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com