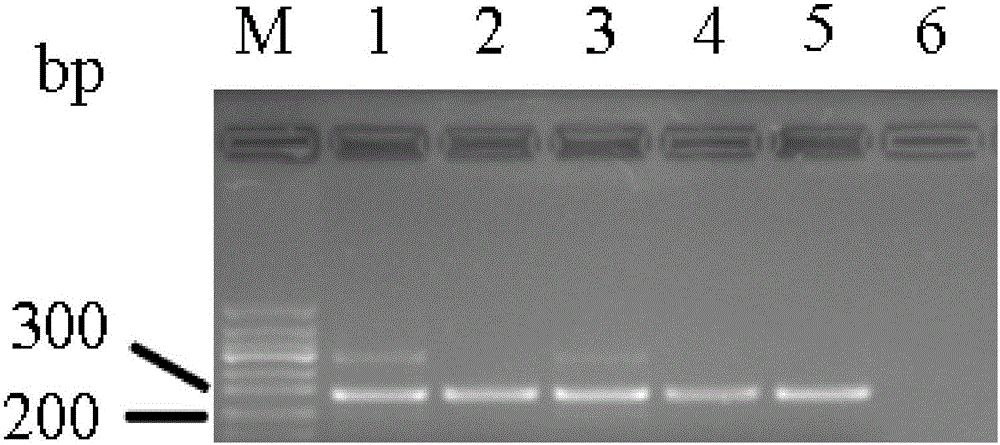
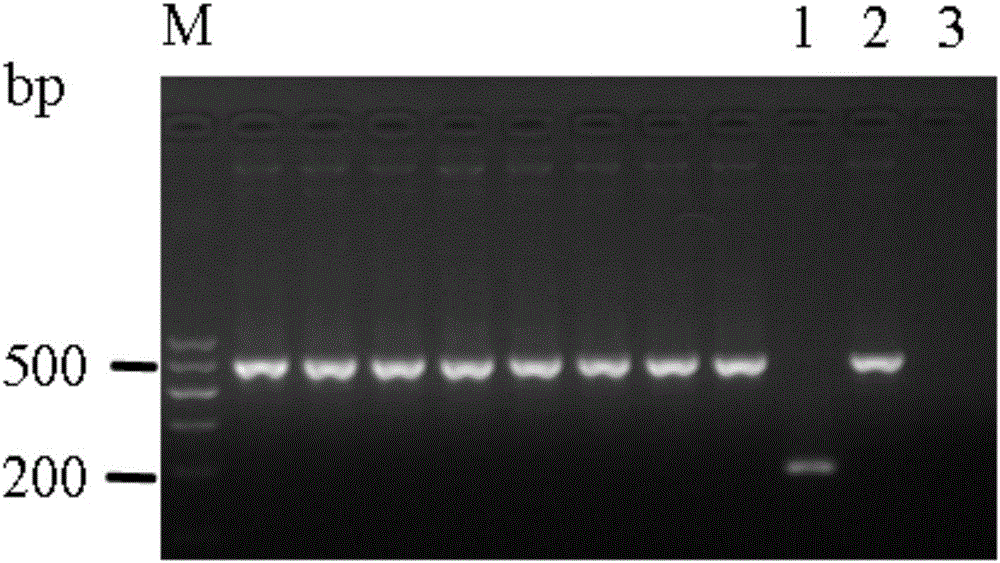
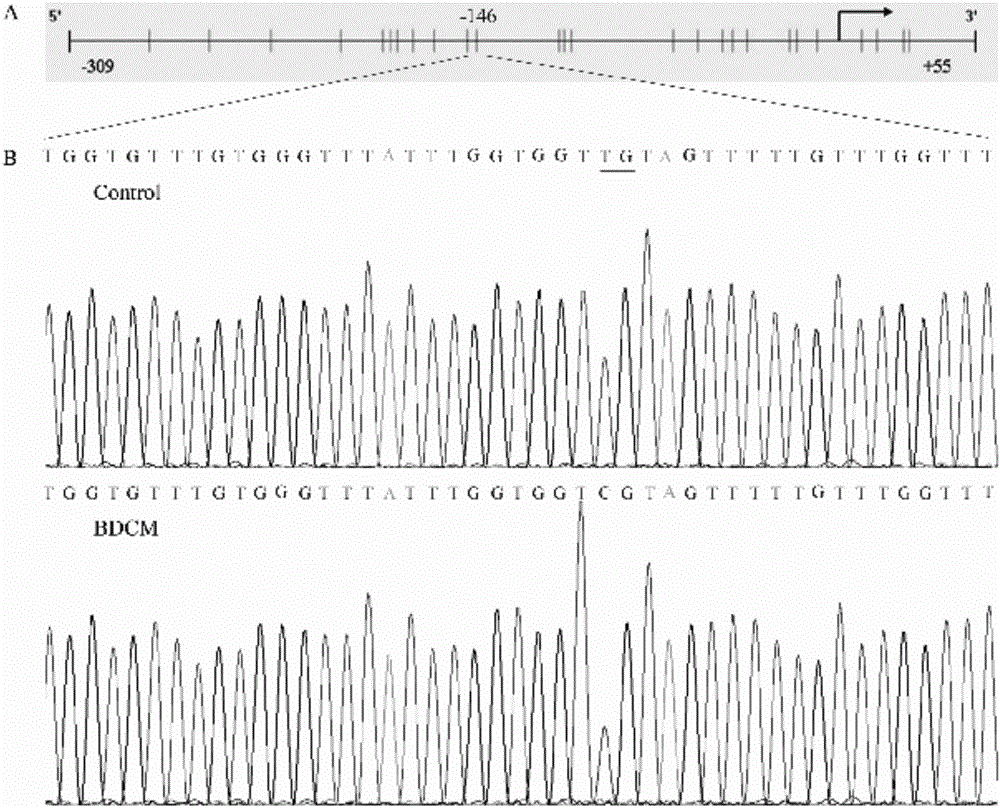
E-cadherin methylation detection method based on hydrosulphite sequencing method
A technology of bisulfite and cadherin, which is applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of high failure rate, improve the success rate, improve specificity and sensitivity, and reduce non-specific The effect of heterosexual amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0025] Embodiments of the invention are described in detail below, but the invention can be practiced in many different ways as defined and covered by the claims.
[0026] Since DNA methylation is closely related to the occurrence and development of tumors, the detection of abnormal DNA methylation sites of specific genes is one of the hot spots in the research of tumor biomarkers. After the genomic DNA to be tested is converted by bisulfite, unmethylated cytosine is converted to uracil, while 5-methylated cytosine remains unchanged. That is to say, the vast majority of methylation detection methods convert the subtle changes of chemical molecules before and after modification into base changes and display them visually through PCR amplification. However, excess genomic DNA is likely to cause incomplete bisulfite modification, resulting in the inability of partially unmethylated cytosine to be effectively converted into uracil, resulting in false positive results.
[0027] Af...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com