Amplification method for 3'-RACE adaptor primer and 3'-end unknown gene sequences
A linker primer and -RACE technology, which is applied in biochemical equipment and methods, microbial measurement/inspection, DNA/RNA fragments, etc., can solve the problems of difficult cloning, interference with target bands, and affecting the amplification efficiency of target fragments, etc. , to achieve the effect of strong pertinence and loss avoidance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] In this embodiment, the complete amplification of the unknown gene sequence at the 3' end is carried out according to the known 5' end sequence of the flounder sox2 gene shown in SEQ ID NO.4, and the specific steps are as follows:
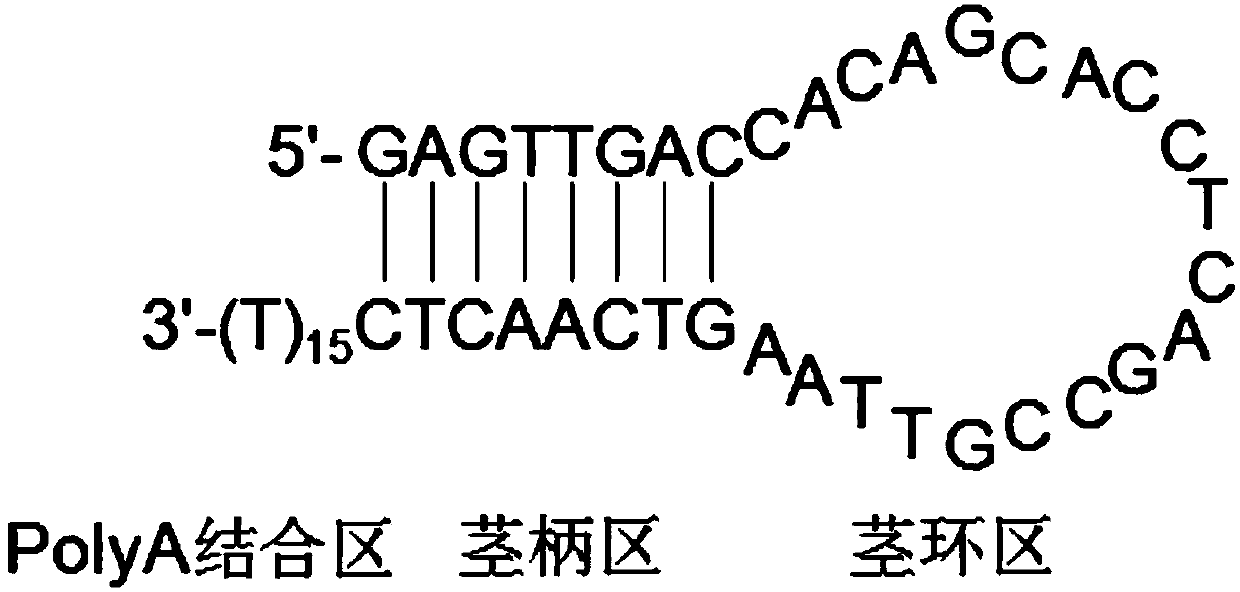
[0042] (1) Design a 3'-RACE linker primer according to the target species and target gene, which has the nucleotide sequence shown in SEQ ID NO.5: 5'-GAGTTGACCACAGCACCTCAGCCGTTAAGTCAACTC(T) 15 -3′, such as figure 1 As shown, the linker primer has a stem-loop structure, consisting of a stem region, a stem-loop region, and a PolyA binding region.
[0043] (2) An anchor primer matching the 3'-RACE linker primer for the amplification of the unknown gene sequence at the 3' end of the flounder sox2 gene and a PCR primer involved in the amplification of the unknown gene sequence at the 3' end were simultaneously designed.
[0044]The anchor primers include 3'-RACE outer primers and 3'-RACE inner primers;
[0045] PCR primers include upstream prim...
Embodiment 2
[0060] The difference of this embodiment is that in step (4), the PCR product obtained in step (3) was diluted 100 times as a template to carry out nested PCR reaction.
[0061] All the other steps are the same as in Example 1.
Embodiment 3
[0063] The difference in this embodiment is that the length of the anchor primer designed in step (2) can vary, and its length can be between 20 and 25 bp, and the 3' end of the 3'-RACE outer primer and the 3'-RACE inner The overlapping sequence at the 5' end of the primer can be between 8 and 12 bp.
[0064] All the other steps are the same as in Example 1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com