Application of miR-489-3p to preparation of medicines for diagnosing and treating human osteoporosis
A technology for osteoporosis and osteoporosis, applied in the fields of molecular biology and medicine, can solve problems not mentioned
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1: Detection of low expression of miR-489-3p in bone tissue samples of senile osteoporosis patients by using human miRNA expression profiling chip and qPCR technology.
[0028] (1) Design and prepare specific primer hsa-miR-489-3p, the sequence is shown in Table 1;
[0029] (2) Collection of bone tissue samples from senile osteoporosis patients:
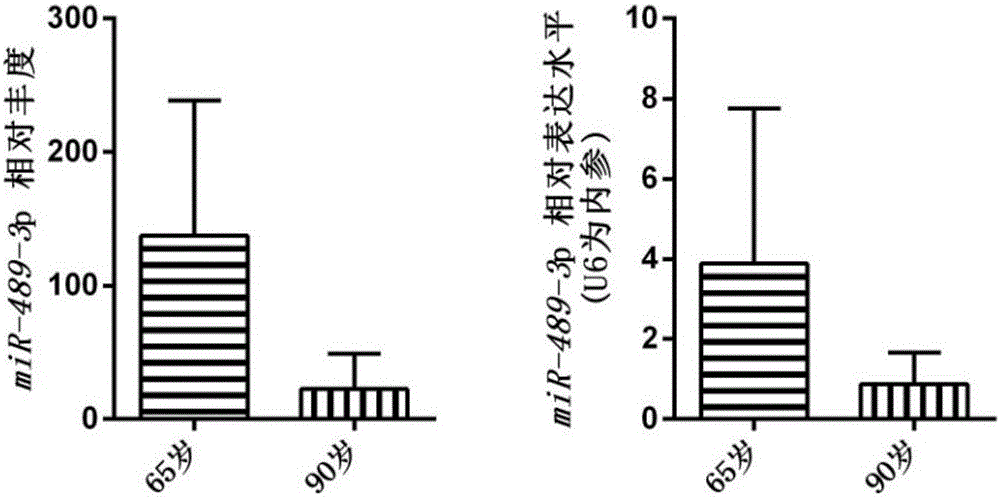
[0030] A total of 111 bone tissue samples from senile osteoporosis patients were collected, immediately preserved in RNA protective agent, and frozen in a -80°C refrigerator. Combined with the patient's past medical history, treatment history and other information, the bone tissue samples of patients without other serious chronic diseases were screened and divided into 90-year-old group and 65-year-old group.
[0031] (3) Extract RNA from bone tissue samples of patients with senile osteoporosis:
[0032] Take the bone tissue sample, peel off the muscle, grind the bone tissue with liquid nitrogen, transfer it to a 1.5ml...
Embodiment 2
[0039] Example 2: Using qPCR technology to detect the low expression of miR-489-3p in the bone tissue of an aged osteoporosis mouse model.
[0040] (1) Design and prepare specific primer mmu-miR-489-3p, the sequence is shown in Table 2;
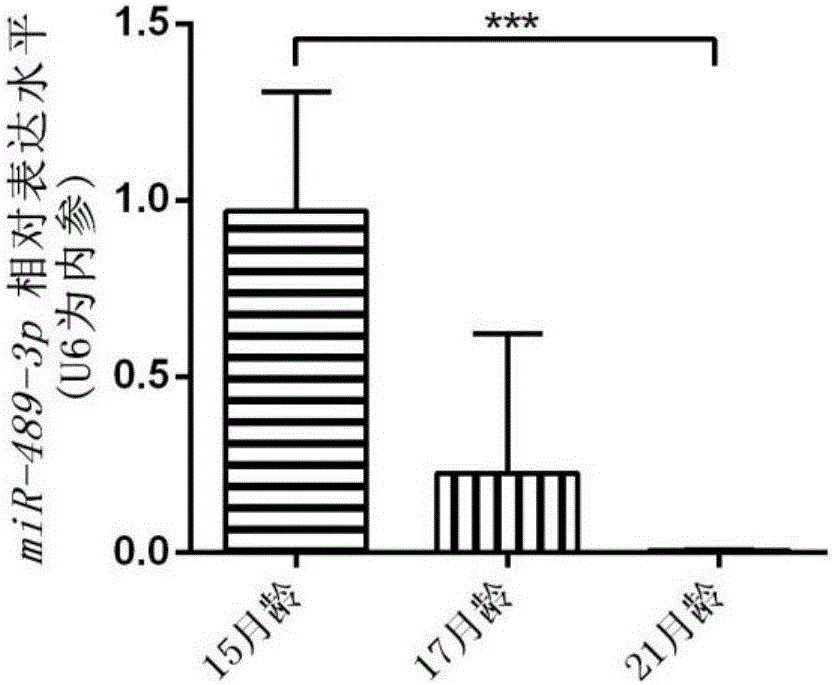
[0041] (2) Establish an aging mouse model of osteoporosis:
[0042] C57BL / 6 experimental mice were selected and raised to 15 months, 17 months, and 21 months. The mice were sacrificed, tibia samples were collected, and stored in a -80°C refrigerator.
[0043] (3) RNA was extracted from the mouse tibia sample, and the expression level of miR-489-3p in the bone tissue of the mouse model was detected by qPCR technology (the steps were the same as above). The sequence of the mmu-miR-489-3p primer used is as follows:
[0044] Table 2 mouse miR-489-3p primer sequence
[0045]
[0046] refer to figure 2 , the results showed that miR-489-3p was low expressed in the bone tissue of aged osteoporosis mouse model, further indicating that miR-489-...
Embodiment 3
[0047] Example 3: Using qPCR technology to detect the low expression of miR-489-3p in the bone tissue of the HLU hindlimb unloaded osteoporosis mouse model.
[0048] (1) Establish HLU hindlimb unloading osteoporosis mouse model:
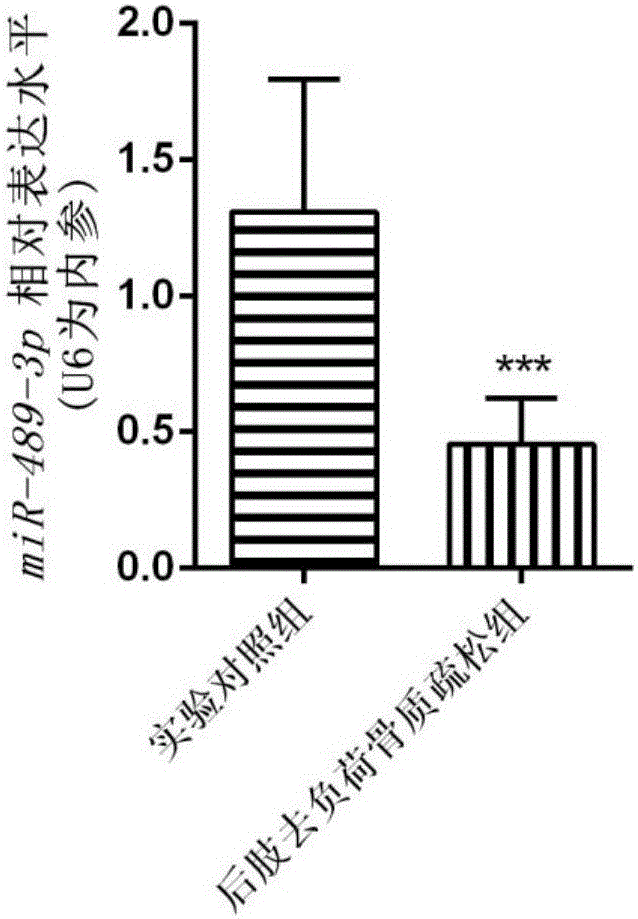
[0049] The C57BL / 6 experimental mice were selected, and the mice were hung on the rings of the tail suspension device with a paper clip, and the mouse spine was 30° from the ground to carry out tail suspension and hindlimb unloading. During the experiment, the animals had free access to food and water, and the mice were sacrificed 28 days later.
[0050] (2) RNA was extracted from the mouse tibia sample, and the expression level of miR-489-3p in the bone tissue of the HLU hindlimb unloaded osteoporosis mouse model was detected by qPCR technology (the steps were the same as above).
[0051] refer to image 3 , the results showed that miR-489-3p was lowly expressed in the bone tissue of the HLU hindlimb unloaded osteoporosis mouse model, which fully ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com